RAST Tutorial
The RAST Server
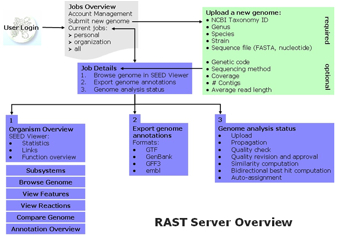
The RAST (Rapid Annotation using Subsystem Technology) Server provides high quality genome annotations for prokaryotes across the whole phylogenetic tree. It makes a SEED-quality annotation available as a service with a 48 hour turnaround time. The SEED environment and SEED data structures (most prominently FIGfams) are used to compute the automatic annotations; however data is not added into the SEED automatically. Once annotation is completed, genomes can be downloaded in a variety of formats or viewed online. The genome annotation provided does include a mapping of genes to subsystems and a metabolic reconstruction. Figure 1 provides an overview of the RAST Server and connections to the SEED Viewer.
Getting Started: Registration is required for genome submission and viewing of results. This enables us to contact users once the computation is finished and in case the users intervention is required.
Figure 1. Overview of the RAST Server navigation, features and capabilities.
Jobs Overview
Upon logging onto the server, users are directed to the “Jobs Overview” page, which as the name suggests, provides a site for job management. Jobs Overview has two main components: starting a new job and reviewing submitted/completed jobs.
Start a new job. The navigation bar (Figure 2) at the top of the page provides a pull down menu for job submission, logout, and review/edit user account information. To start a new job, users should select “Upload Genome” from the navigation bar or the link near the top of the page. The user is required to provide a valid taxonomy id+, the organism’s Genus, species, and strain, as well as a nucleotide sequence file in FASTA format. Optional parameters are suggested, but not required and include genetic code, sequencing method, coverage, number of contigs and average read length. Currently the server supports genome analysis of prokaryotes with genetic codes 4 and 11.
+Taxonomy id’s can be obtained from the NCBI taxonomy browser (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/). Search by organism name, and the taxonomy id is returned. For example, Escherichia coli K12 has taxonomy id 83333.
Figure 2. Jobs Overview Navigation Bar.
Reviewing submitted/completed jobs.
The overall status of genome analysis can be viewed from the main portion of the Jobs Overview page. This contains information regarding a user’s personal jobs and when applicable, jobs of the user’s organization. Figure 3 shows an example account where the individual does not have any personal jobs, but has access to several for their organization. The table shows each job/genome and its status and contains information including job number, name of the user who started the job, genome id (taxonomy_id. internal_id), genome name, and annotation progress.
The table of jobs can be sorted on any column containing textual information. When the user has numerous genomes to select from, they can use the text boxes in the table header to search and refine the list of jobs.
Clicking on the bars for a given job in the annotation progress column directs the user to the “Job Details” page where the detailed job status and access to the genome analysis can be found.
Figure 3. Jobs Overview for a given account.
Job Details
For a given job, the Job Details page provides the user with information regarding the status of the genome annotation progress, as well as access to the results of the analysis upon completion.
Account and job management links are found in the navigation bar at the top of the Job Details page (Figure 4) includes (1) logout, (2) upload a new genome, (3) link back to the Jobs Overview, and (4) review/edit your account information.
Figure 4. Job Details Navigation Bar.
The Job Details page has three main functions: 1. To provide access to the results of the genome analysis via the SEED Viewer, 2. Export tool that enables the user to download the annotated genome in various formats (GTF, GenBank, GFF3, or EMBL) 3. The status of their genome analysis. Information regarding the status of each major step in the analysis process is reported which includes: o Genome upload o Genome id and Name o Job number o Name of user who created the job o Date and time of job submission o Rapid propagation (protein function annotation) o Quality check o Statistics (number of features, warnings, fatal problems) o Warnings (overlaps) o Fatal Problems (embedded genes) o Quality revision (users approval) o Similarity Computation o Bidirectional Best Hit Computation (for conserved regions and functional coupling) o Auto Assignment (to subsystems)