Difference between revisions of "MG RAST Tutorial"
RobEdwards (talk | contribs) |
RobEdwards (talk | contribs) |
||
| Line 53: | Line 53: | ||
The metagenomics-RAST is primarily designed to handle random community genomes. At the moment, we only provide rudimentary support for 16S DNA sequence analysis, although this is near the very top of our to-do list. | The metagenomics-RAST is primarily designed to handle random community genomes. At the moment, we only provide rudimentary support for 16S DNA sequence analysis, although this is near the very top of our to-do list. | ||
| − | Our colleagues at San Diego State University have developed two different tools for handling 16S rDNA sequences. FastGroup, a stand-alone java application ([http://www.ncbi.nlm.nih.gov/pubmed/11707150?ordinalpos=2&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Seguritan V and Rohwer F. (2001) FastGroup: a program to dereplicate libraries of 16S rDNA sequences]. BMC Bioinformatics. 2:9. Epub 2001 Oct 16.) is the original program, and it was updated to FastGroupII ([http://www.ncbi.nlm.nih.gov/pubmed/16464253?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Yu Y, Breitbart M, McNairnie P, and Rohwer F. (2006) FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries]. BMC Bioinformatics. Feb 7;7:57.). We have provided some [[instructions for using FastGroupII with large data sets]]. | + | Our colleagues at San Diego State University have developed two different tools for handling 16S rDNA sequences. FastGroup, a stand-alone java application ([http://www.ncbi.nlm.nih.gov/pubmed/11707150?ordinalpos=2&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Seguritan V and Rohwer F. (2001) FastGroup: a program to dereplicate libraries of 16S rDNA sequences]. BMC Bioinformatics. 2:9. Epub 2001 Oct 16.) is the original program, and it was updated to FastGroupII ([http://www.ncbi.nlm.nih.gov/pubmed/16464253?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Yu Y, Breitbart M, McNairnie P, and Rohwer F. (2006) FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries]. BMC Bioinformatics. Feb 7;7:57.). We have provided some [[instructions for using FastGroupII with large data sets]]. We recommend FastGroupII for clustering and primary analysis of 16S libraries, and then the data from that can be fed into RDP Classifier and other programs. |
Revision as of 21:17, 23 March 2008
Overview
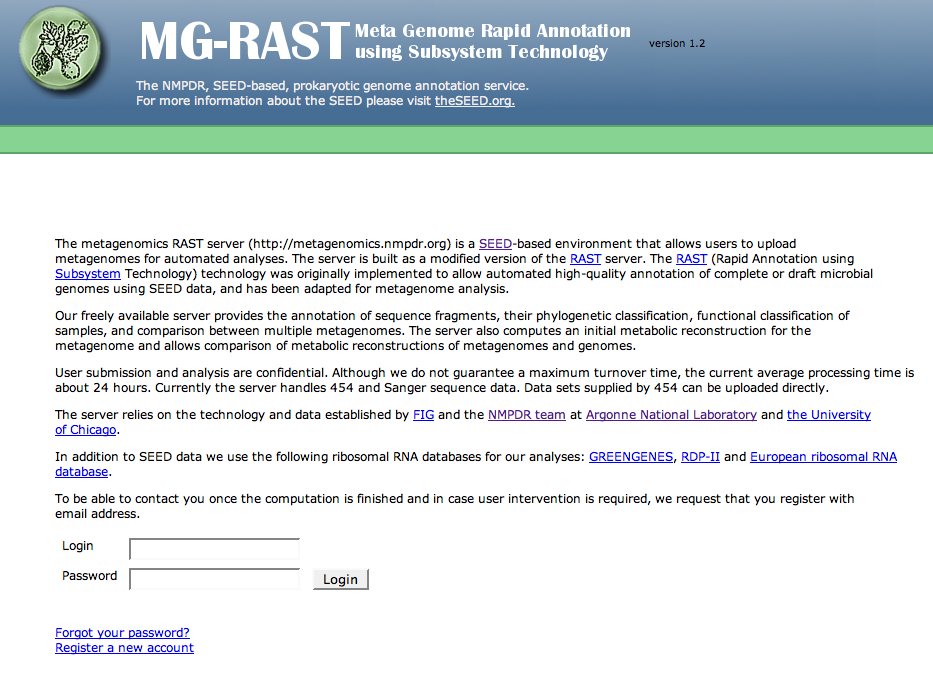
The metagenomics RAST server (http://metagenomics.nmpdr.org) is a SEED-based environment that allows users to upload metagenomes for automated analyses. The server is built as a modified version of the RAST server. The RAST (Rapid Annotation using Subsystem Technology) technology was originally implemented to allow automated high-quality annotation of complete or draft microbial genomes using SEED data, and has been adapted for metagenome analysis.
Our freely available server provides the annotation of sequence fragments, their phylogenetic classification, functional classification of samples, and comparison between multiple metagenomes. The server also computes an initial metabolic reconstruction for the metagenome and allows comparison of metabolic reconstructions of metagenomes and genomes.
User submission and analysis are confidential. Although we do not guarantee a maximum turnover time, the current average processing time is about 24 hours. Currently the server handles 454 and Sanger sequence data. Data sets supplied by 454 can be uploaded directly. In either case, the data needs to be in Valid fasta format. For more information, please see Which Sequences Should I Upload, and Where. For the metagenomics service please also read this explanation of metagenomics sequence formats.
The server relies on the technology and data established by FIG and the NMPDR team at Argonne National Laboratory and the University of Chicago.
In addition to SEED data we use the following ribosomal RNA databases for our analyses: greengenes, RDP-II and European ribosomal RNA database.
Registration
Registration is required for metagenome submission and viewing of results. This enables us to contact users once the computation is finished and in case the users intervention is required.
At the bottom of the main page is a like for registration (see Figure 1).
Required fields for registration include first and last name and your valid email address. Login information and other communication regarding the status of your metagenome analysis job(s) will be sent to the email address you provide. Optional information includes your organization and any notes you would like to send the rast server support team.
Please note that your login and password are valid for use in both the MG-RAST and RAST servers.
Submitting a Job
Once you have registered and logged into the server, you will be directed to your Jobs Overview. At the top of this page will be a link labeled "Upload Genome" which will allow you to start a new job.
Your metagenome file(s) should be uploaded as either a single plain text file containing all the sequences in FASTA format, or a gzip compressed tar archive (tar.gz) that has your FASTA sequences.
Please do not upload uncompressed files larger than 30 MB. If your data set is larger, use the compressed format or contact us for other options. If you would like, you can also include the quality files in your archive. The fasta file names should end either *.fna, *.fa, or *.fasta, and the quality files should be named *.qual. The quality files are not currently used in the analysis, but the sequences will be renamed and renumbered along with the fasta sequences. If you have trouble with the upload format please email [email protected] and we'll be happy to help.
Data entered into the server will not be used for any purposes or integrated into the main SEED environment, it will remain on this server for 120 days or until deleted by the submitting user.
An email will be sent once the automatic annotation has finished or in case user intervention is required.
Viewing Results
The overall status of your metagenome analyses can be viewed from the main portion of the Jobs Overview page. This contains information regarding a user’s personal jobs and when applicable, jobs of the user’s organization. Information includes each job/metagenome and its status and contains information including job number, name of the user who started the job, metagenome name, and annotation progress.
The table of jobs can be sorted on any column containing textual information. When the user has numerous metagenomes to select from, they can use the text boxes in the table header to search and refine the list of jobs.
Clicking on the bars for a given job in the annotation progress column directs the user to the “Job Details” page where the detailed job status and access to the metagenome analysis can be found ("Browse annotated genome in SEED Viewer"). Users can also download the results in compressed GenBank format.
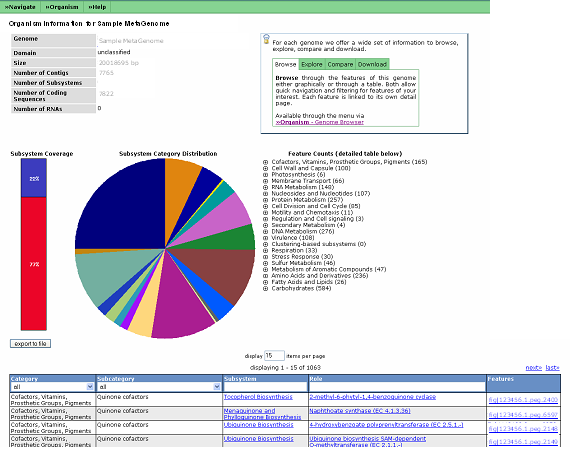
MetaGenome Overview
The MetaGenome Overview provides the user with various statistics regarding their metagenome and details on how each of these numbers are calculated can be found here.
Users can search for a given function, subsystem or process in the table, or browse the Subsystem Overview. At the top right hand side of the page is a set of tabs that offer a wide set of information to browse, explore, compare and download. Browse allows users to look through the features of this metagenome either graphically or through a table. Both allow quick navigation and filtering for features of your interest. Each feature is linked to its own detail page. Explore allows users to view scenarios. Scenarios are isolated metabolic divisions that in aggregate represent the metabolic functionality of the metagenome. Each scenario is tested for reaction availability against the annotated functions. They provide the foundation for generating a metabolic reconstruction. Comparison of two metagenomes is also possible via the compare tab. You can also export all information about this metagenome (e.g. annotations, scenarios, subsystems) into a variety of formats (e.g. EMBL, Excel) for further analysis on your own system.
16S Sequences
The metagenomics-RAST is primarily designed to handle random community genomes. At the moment, we only provide rudimentary support for 16S DNA sequence analysis, although this is near the very top of our to-do list.
Our colleagues at San Diego State University have developed two different tools for handling 16S rDNA sequences. FastGroup, a stand-alone java application (Seguritan V and Rohwer F. (2001) FastGroup: a program to dereplicate libraries of 16S rDNA sequences. BMC Bioinformatics. 2:9. Epub 2001 Oct 16.) is the original program, and it was updated to FastGroupII (Yu Y, Breitbart M, McNairnie P, and Rohwer F. (2006) FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics. Feb 7;7:57.). We have provided some instructions for using FastGroupII with large data sets. We recommend FastGroupII for clustering and primary analysis of 16S libraries, and then the data from that can be fed into RDP Classifier and other programs.