The Next Step: Formulating State Machine Models for Microbes:
by Ross Overbeek
October, 2010
In this document, I plan to go through the following steps:
- First, I will go through some examples of conjectures that follow naturally from a minimal
study of atomic regulons (sets of genes that are always co-expressed).
My goal is to show that a very simple integration of data (relating to operons, subsystems, and expression data)
produces truly impressive results. I have recently collected well over a hundred examples of atomic
regulons that can be easily used to produce testable conjectures.
- Then, I will reflect on why these conjectures are easy to produce.
- I will argue that they are just a by-product of a more important issue -- the need to construct state-machine
models based on atomic regulons.
- Finally, I will describe what I see as the pipeline needed to really exploit atomic regulons.
Some Examples in the Use of Atomic Regulons
An Example Relating to Ubiquinone/Menaquinone Metablism
As my first example of what might be achievable by studying the atomic regulaons,
contider Atomic Regulon 14 in our current set for E.coli:
Pegs in Atomic Regulon 14 [ON=888 OFF=6]
Pearson Coefficients:
| PEG |
peg.2858 |
peg.2859 |
peg.2860 |
peg.2861 |
peg.3759 |
peg.3760 |
peg.3761 |
peg.3762 |
peg.3763 |
peg.3764 |
peg.3765 |
peg.3769 |
peg.3770 |
| fig|83333.1.peg.2858 |
|
0.793 |
0.783 |
0.707 |
0.436 |
0.676 |
0.659 |
0.610 |
0.599 |
0.692 |
0.565 |
0.700 |
0.701 |
| fig|83333.1.peg.2859 |
0.793 |
|
0.861 |
0.740 |
0.462 |
0.743 |
0.701 |
0.570 |
0.667 |
0.696 |
0.590 |
0.736 |
0.707 |
| fig|83333.1.peg.2860 |
0.783 |
0.861 |
|
0.850 |
0.437 |
0.735 |
0.668 |
0.547 |
0.632 |
0.683 |
0.468 |
0.727 |
0.714 |
| fig|83333.1.peg.2861 |
0.707 |
0.740 |
0.850 |
|
0.505 |
0.758 |
0.741 |
0.665 |
0.731 |
0.727 |
0.484 |
0.727 |
0.697 |
| fig|83333.1.peg.3759 |
0.436 |
0.462 |
0.437 |
0.505 |
|
0.721 |
0.802 |
0.766 |
0.531 |
0.613 |
0.517 |
0.634 |
0.540 |
| fig|83333.1.peg.3760 |
0.676 |
0.743 |
0.735 |
0.758 |
0.721 |
|
0.899 |
0.867 |
0.856 |
0.857 |
0.761 |
0.818 |
0.813 |
| fig|83333.1.peg.3761 |
0.659 |
0.701 |
0.668 |
0.741 |
0.802 |
0.899 |
|
0.852 |
0.740 |
0.853 |
0.690 |
0.817 |
0.801 |
| fig|83333.1.peg.3762 |
0.610 |
0.570 |
0.547 |
0.665 |
0.766 |
0.867 |
0.852 |
|
0.816 |
0.844 |
0.778 |
0.717 |
0.728 |
| fig|83333.1.peg.3763 |
0.599 |
0.667 |
0.632 |
0.731 |
0.531 |
0.856 |
0.740 |
0.816 |
|
0.800 |
0.749 |
0.736 |
0.731 |
| fig|83333.1.peg.3764 |
0.692 |
0.696 |
0.683 |
0.727 |
0.613 |
0.857 |
0.853 |
0.844 |
0.800 |
|
0.769 |
0.786 |
0.794 |
| fig|83333.1.peg.3765 |
0.565 |
0.590 |
0.468 |
0.484 |
0.517 |
0.761 |
0.690 |
0.778 |
0.749 |
0.769 |
|
0.573 |
0.661 |
| fig|83333.1.peg.3769 |
0.700 |
0.736 |
0.727 |
0.727 |
0.634 |
0.818 |
0.817 |
0.717 |
0.736 |
0.786 |
0.573 |
|
0.891 |
| fig|83333.1.peg.3770 |
0.701 |
0.707 |
0.714 |
0.697 |
0.540 |
0.813 |
0.801 |
0.728 |
0.731 |
0.794 |
0.661 |
0.891 |
|
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.2858 |
2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase (EC 1.14.13.-) |
CBSS-87626.3.peg.3639,Ubiquinone_Biosynthesis |
| fig|83333.1.peg.2859 |
2-octaprenyl-6-methoxyphenol hydroxylase (EC 1.14.13.-) |
CBSS-87626.3.peg.3639,Ubiquinone_Biosynthesis |
| fig|83333.1.peg.2860 |
Xaa-Pro aminopeptidase (EC 3.4.11.9) |
CBSS-87626.3.peg.3639 |
| fig|83333.1.peg.2861 |
FIG037488: Putative conserved exported protein precursor |
CBSS-87626.3.peg.3639 |
| fig|83333.1.peg.3759 |
DNA recombination protein RmuC |
DNA_repair,_bacterial |
| fig|83333.1.peg.3760 |
Ubiquinone/menaquinone biosynthesis methyltransferase UbiE (EC 2.1.1.-) |
Menaquinone_Biosynthesis_via_Futalosine,Menaquinone_and_Phylloquinone_Biosynthesis,Ubiquinone_Biosynthesis |
| fig|83333.1.peg.3761 |
Protein YigP (COG3165) clustered with ubiquinone biosynthetic genes |
Menaquinone_Biosynthesis_via_Futalosine,Ubiquinone_Biosynthesis |
| fig|83333.1.peg.3762 |
Ubiquinone biosynthesis monooxygenase UbiB |
Ubiquinone_Biosynthesis |
| fig|83333.1.peg.3763 |
Twin-arginine translocation protein TatA |
Cluster-based_Subsystem_Grouping_Hypotheticals_-_perhaps_Proteosome_Related,Twin-arginine_translocation_system,YgfZ |
| fig|83333.1.peg.3764 |
Twin-arginine translocation protein TatB |
Twin-arginine_translocation_system |
| fig|83333.1.peg.3765 |
Twin-arginine translocation protein TatC |
Cluster-based_Subsystem_Grouping_Hypotheticals_-_perhaps_Proteosome_Related,Twin-arginine_translocation_system |
| fig|83333.1.peg.3769 |
3-polyprenyl-4-hydroxybenzoate carboxy-lyase (EC 4.1.1.-) |
Menaquinone_Biosynthesis_via_Futalosine,Ubiquinone_Biosynthesis |
| fig|83333.1.peg.3770 |
NAD(P)H-flavin reductase (EC 1.5.1.29) (EC 1.16.1.3) |
Ubiquinone_Biosynthesis |
This is a fairly big "atomic regulon", and it may well not really be "atomic". What we see, it seems to me, is two large gene clusters that seem
to have very similar expression profiles. One goes from PEG fig|83333.1.peg.2858 to PEG fig|83333.1.peg.2861. The other goes from
PEG fig|83333.1.peg.3759 through PEG fig|83333.1.peg.3770.
The first cluster appears to include two genes relating to ubiquinone/menaquinone metabolism, as well as two
less characterized genes. The second cluster appears to include four genes relating to ubiquinone/menaquinone metabolism,
three genes relating to a twin-arginine translocation system, a DNA repair gene (rmuC) and a hypothetical (fig|83333.1.peg.3761).
There are a number of relevant points that we should ponder:
- First, let us note the expression profiles. It appears that each of the clusters was expressed in 888 experiments and not expressed in 6.
It would be good to know what those six experiments were, I think.
- The fact that significant genes from the same metabolic activity (ubiquinone/menaquinone metabolism) are present in both
clusters supports the position that the clusters are actually related.
- It needs to be noted that, while the first cluster may be an operon, the second is not. The gene
fig|83333.1.peg.3768 is a transcriptional regulator that breaks the cluster into
two potential operons (each containing two of the ubiquinone/menaquinone genes).
The obvious conjectures would be as follows:
-
The PEG fig|83333.1.peg.3761 is embedded between ubiquinone/menaquinone gene in a single operon.
It is clearly a good candidate for a role in ubiquinone/menaquinone metabolism.
-
A similar comment could be made for the tatABC genes. The Tat
(twin-arginine translocation) system mediates export of periplasmic
proteins in folded conformation. It would take an expert to
understand how this might relate to the metabolism in question, if at
all. However, it does seem to me that both processes (the electron transport
and movement of proteins across a membrane) might be related somehow.
The Twin-Arginine Transport System
Frank Sargent, Ben C. Berks and Tracy Palmer
The twin-arginine transport (Tat) system is a protein-targeting
pathway found in the cytoplasmic membranes of many eubacteria, some
archaea, and the chloroplasts and mitochondria of plants. It is
apparently not a feature of animal physiology. Substrate proteins are
targeted to a membrane-bound transport apparatus by N-terminal signal
peptides harbouring a distinctive ‘twin-arginine’ amino acid sequence
motif, and, most remarkably, all substrate proteins are transported in
a fully folded conformation. Model systems most commonly used to study
the fundamentals of Tat transport are the Gram-negative eubacterium
Escherichia coli, the Gram-positive eubacterium Bacillus subtilis, and
thylakoid membranes derived from pea or maize chloroplasts.
- The RmuC protein is not terribly well characterized. The sole paper that I could find
was
Genes Cells. 2000 Jun;5(6):425-37.
Genes involved in the determination of the rate of inversions at short inverted repeats.
Slupska MM, Chiang JH, Luther WM, Stewart JL, Amii L, Conrad A, Miller JH.
Abstract
BACKGROUND: Not all of the enzymatic pathways involved in genetic
rearrangements have been elucidated. While some rearrangements occur
by recombination at areas of high homology, others are mediated by
short, often interrupted homologies. We have previously constructed an
Escherichia coli strain that allows us to examine inversions at
microhomologies, and have shown that inversions can occur at short
inverted repeats in a recB,C-dependent fashion.
RESULTS: Here, we report on the use of this strain to define genetic
loci involved in limiting rearrangements on an F' plasmid carrying the
lac genes. Employing mini-Tn10 derivatives to generate insertions near
or into genes of interest, we detected three loci (rmuA,B,C) that,
when mutated, increase inversions. We have mapped, cloned and
sequenced these mutator loci. In one case, inactivation of the sbcC
gene leads to an increase in rearrangements, and in another,
insertions near the recE gene lead to an even larger increase. The
third gene involved in limiting inversions, rmuC, has been mapped at
86 min on the E. coli chromosome and encodes a protein of unknown
function with a limited homology to myosins, and some of the SMC
(structural maintenance of chromosomes) proteins.
CONCLUSIONS: This work presents the first example of an anti-mutator
role of the sbcC,D genes, and defines a new gene (rmuC) involved in
DNA recombination.
Here is a description of a PFAM that matches the gene:
RmuC family
This family contains several bacterial RmuC DNA recombination
proteins. The function of the RMUC protein is unknown but it is
suspected that it is either a structural protein that protects DNA
against nuclease action, or is itself involved in DNA cleavage at the
regions of DNA secondary structures.
There is a puzzle here relating to electron transport,
transportation across a network (of folded proteins?),
and structural aspects of DNA.
I think that we need to study the six experiments, focus on the
easiest question now, and think of how to gain more data. I am not
sure what it would take to implicate the 7 genes of interest (PEGs
2860, 2861, 3759, 3761, 3763, 3764, and 3765) in ubiquinone metabolism and to clarify their role.
An Example of a Hypothetical Protein Implicated in the TCA: E.coli Atomic Regulon 14
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.713 |
Succinate dehydrogenase cytochrome b-556 subunit |
Succinate_dehydrogenase |
| fig|83333.1.peg.714 |
Succinate dehydrogenase hydrophobic membrane anchor protein |
Succinate_dehydrogenase |
| fig|83333.1.peg.715 |
Succinate dehydrogenase flavoprotein subunit (EC 1.3.99.1) |
Serine-glyoxylate_cycle,Succinate_dehydrogenase,TCA_Cycle |
| fig|83333.1.peg.716 |
Succinate dehydrogenase iron-sulfur protein (EC 1.3.99.1) |
5-FCL-like_protein,Serine-glyoxylate_cycle,Succinate_dehydrogenase,TCA_Cycle,YgfZ |
| fig|83333.1.peg.717 |
hypothetical protein |
|
| fig|83333.1.peg.718 |
2-oxoglutarate dehydrogenase E1 component (EC 1.2.4.2) |
Dehydrogenase_complexes,TCA_Cycle |
| fig|83333.1.peg.719 |
Dihydrolipoamide succinyltransferase component (E2) of 2-oxoglutarate dehydrogenase complex (EC 2.3.1.61) |
Dehydrogenase_complexes,Lipoic_acid_metabolism,TCA_Cycle |
Let me begin by saying that it is not clear that this is a real gene, although numerous groups
have called it. It is very short (86 aa). If it is real, I claim that it is related to the TCA,
and probably to the succinate dehydrogenase or the 2-oxoglutarate dehydrogenase. As I understand the situation, the
expression data is overwhelming support that the gene is expressed and translated.
The expression data that we have for this atomic regulon suggests that
it was ON in approximately 840 experiments and OFF in about 50-60. We can graph the expression levels
for genes 716,717,and 718 to see how they relate, and the correspondence looks pretty good to me:
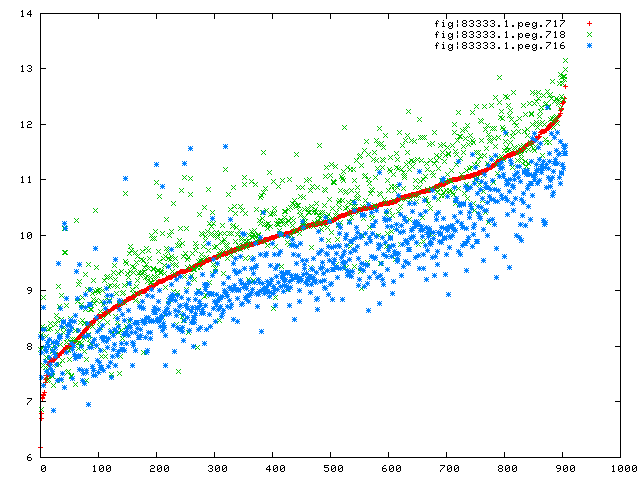
Here, we have sorted the experiments based on the expression levels of 717 (so the lowest values
on the X-axis are for experiments in which 717 had the smallest values), and the expression levels for
the 3 genes are shown with the Y-axis being the normalized values.
An Example Relating to Heme/Siroheme Biosynthesis: E.coli Atomic Regulon 16 [ON=748 OFF=115]
Here is another somewhat complex atomic regulon. Note:
- The genes are from four distinct areas of the genome. Gene 232 is not in a cluster, 3727-3730 form a cluster,
3773-3776 form a cluster, and 3910-3912 form a cluster.
- Three of the four groups relate to Heme and Siroheme Biosynthesis.
- There are two peptidases -- one in the singleton group and one in a group containing a component
of heme/siroheme biosynthesis.
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.232 |
Aminoacyl-histidine dipeptidase (Peptidase D) (EC 3.4.13.3) |
Recycling_of_Peptidoglycan_Amino_Acids |
| fig|83333.1.peg.3727 |
Homolog of E. coli HemY protein |
Experimental_tye,Heme_and_Siroheme_Biosynthesis |
| fig|83333.1.peg.3728 |
Homolog of E. coli HemX protein |
Dissimilatory_nitrite_reductase,Heme_and_Siroheme_Biosynthesis |
| fig|83333.1.peg.3729 |
Uroporphyrinogen-III synthase (EC 4.2.1.75) |
Experimental_tye,Heme_and_Siroheme_Biosynthesis |
| fig|83333.1.peg.3730 |
Porphobilinogen deaminase (EC 2.5.1.61) |
Experimental_tye,Heme_and_Siroheme_Biosynthesis |
| fig|83333.1.peg.3773 |
Xaa-Pro dipeptidase PepQ (EC 3.4.13.9) |
|
| fig|83333.1.peg.3774 |
hypothetical conserved protein COG1739 |
Experimental_tye |
| fig|83333.1.peg.3775 |
Potassium uptake protein TrkH |
Potassium_homeostasis |
| fig|83333.1.peg.3776 |
Protoporphyrinogen IX oxidase, oxygen-independent, HemG (EC 1.3.-.-) |
Experimental_tye,Heme_and_Siroheme_Biosynthesis |
| fig|83333.1.peg.3910 |
Uroporphyrinogen III decarboxylase (EC 4.1.1.37) |
Experimental_tye,Heme_and_Siroheme_Biosynthesis |
| fig|83333.1.peg.3911 |
Endonuclease V (EC 3.1.21.7) |
DNA_repair,_bacterial |
| fig|83333.1.peg.3912 |
hypothetical protein |
|
I believe that development of a coherent story for this regulon will require someone with far more understanding
than I of biochemistry and phisiology.
A Membrane-bound Ni,Fe hydrogenase: E.coli Atomic Regulon 19 [ON=206 OFF=695]
Now, let us shift to an atomic regulon that relates to a membrane-bound Ni,Fe-hydrogenase
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.956 |
Uptake hydrogenase small subunit precursor (EC 1.12.99.6) |
Hydrogenases,Membrane-bound_Ni,_Fe-hydrogenase |
| fig|83333.1.peg.957 |
Uptake hydrogenase large subunit (EC 1.12.99.6) |
Hydrogenases,Membrane-bound_Ni,_Fe-hydrogenase |
| fig|83333.1.peg.958 |
Ni,Fe-hydrogenase I cytochrome b subunit |
Hydrogenases,Membrane-bound_Ni,_Fe-hydrogenase |
| fig|83333.1.peg.959 |
Hydrogenase maturation protease (EC 3.4.24.-) |
Hydrogenases,Membrane-bound_Ni,_Fe-hydrogenase |
| fig|83333.1.peg.960 |
Hydrogenase maturation factor hoxO |
Membrane-bound_Ni,_Fe-hydrogenase |
| fig|83333.1.peg.961 |
Hydrogenase maturation factor hoxQ |
Membrane-bound_Ni,_Fe-hydrogenase |
| fig|83333.1.peg.962 |
Cytochrome d ubiquinol oxidase subunit I (EC 1.10.3.-) |
Cytochrome_c_oxidases_d@O_copy,Terminal_cytochrome_d_ubiquinol_oxidases,Terminal_cytochrome_oxidases |
| fig|83333.1.peg.963 |
Cytochrome d ubiquinol oxidase subunit II (EC 1.10.3.-) |
Bacterial_RNA-metabolizing_Zn-dependent_hydrolases,Conserved_gene_cluster_associated_with_Met-tRNA_formyltransferase,Cytochrome_c_oxidases_d@O_copy,Terminal_cytochrome_d_ubiquinol_oxidases,Terminal_cytochrome_oxidases |
| fig|83333.1.peg.964 |
Putative periplasmic protein |
|
| fig|83333.1.peg.965 |
Phosphoanhydride phosphohydrolase (EC 3.1.3.2) (pH 2.5 acid phosphatase) (AP) / 4- phytase (EC 3.1.3.26) |
Capsular_Polysaccharides_Biosynthesis_and_Assembly |
Let me show you the correspondences in expression between the first gene in this long cluster (956) and two
of the genes near the end:
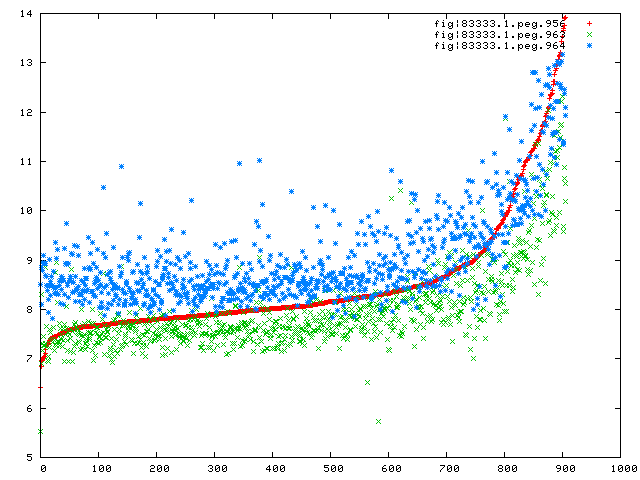
I believe that these tables (and the expression correspondences) mean that the roles of the last four genes in
the cluster (962-965) must be interpreted within their presence in the cluster. I consider the existing
annotations as unreliable suggestions that could be substantially improved.
Guessing the Substrate of a Transport System: Atomic Regulon 72 [ON=790 OFF=59]
We have hundreds of cases in which we have a co-expressed set of genes that make up a transport system.
In some cases, other genes are included in the cluser and they offer substantial clues for
characterizing the substrate of the mechanism. Consider the following five genes:
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.402 |
S-adenosylmethionine:tRNA ribosyltransferase-isomerase (EC 5.-.-.-) |
Queuosine-Archaeosine_Biosynthesis,tRNA_modification_Bacteria |
| fig|83333.1.peg.403 |
tRNA-guanine transglycosylase (EC 2.4.2.29) |
Queuosine-Archaeosine_Biosynthesis,tRNA_modification_Bacteria |
| fig|83333.1.peg.404 |
Preprotein translocase subunit YajC (TC 3.A.5.1.1) |
|
| fig|83333.1.peg.405 |
Protein-export membrane protein SecD (TC 3.A.5.1.1) |
|
| fig|83333.1.peg.406 |
Protein-export membrane protein SecF (TC 3.A.5.1.1) |
|
The genes in the cluster are believed to be ON in 790 experiments and OFF in about 60.
If I go to Wikipedia, I find
Queuosine is a modified nucleoside that is present in certain tRNAs in
bacteria and eukaryotes. Originally identified in E. coli,
queuosine was found to occupy the first anticodon position of tRNAs
for histidine, aspartic acid, asparagine and tyrosine. The first
anticodon position pairs with the third "wobble" position in codons,
and queuosine improves accuracy of translation. Synthesis of
queuosine begins with GTP. In bacteria, two classes of riboswitch are
known to regulate genes that are involved in the synthesis or
transport of pre-queuosine1, a precursor to queuosine: PreQ1-I
riboswitches and PreQ1-II riboswitches.
The transport mechanism seems to be for moving proteins. That would
raise the question "What proteins?". The assertion that queusine can
be synthesized or made from pre-queuosine (transported) leads me to
suggest that we consider the possibility that pre=queuosine is the
substrate. I would not do an experiment before consulting someone who
knows something about tRNA modification, but I believe that it does
illustrate a class of clues quite nicely. I have sent the clue off to
a biologist who might be able to predict the substrate much more
accurately than I could.
A Hypothetical Connected to a Sulfur Relay Mediation Complex: E.coli Atomic Regulon 96 [ON=517 OFF=199]
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.3277 |
tRNA 5-methylaminomethyl-2-thiouridine synthase TusB |
Glutathione-regulated_potassium-efflux_system_and_associated_functions,mnm5U34_biosynthesis_bacteria,tRNA_modification_Bacteria |
| fig|83333.1.peg.3278 |
tRNA 5-methylaminomethyl-2-thiouridine synthase TusC |
CBSS-326442.4.peg.1852,Glutathione-regulated_potassium-efflux_system_and_associated_functions,mnm5U34_biosynthesis_bacteria,tRNA_modification_Bacteria |
| fig|83333.1.peg.3279 |
tRNA 5-methylaminomethyl-2-thiouridine synthase TusD |
CBSS-326442.4.peg.1852,Glutathione-regulated_potassium-efflux_system_and_associated_functions,mnm5U34_biosynthesis_bacteria,tRNA_modification_Bacteria |
| fig|83333.1.peg.3280 |
Putative DNA-binding protein |
|
Here we have three genes that form a complex: TusBCD. Two papers have been written about this complex in the last few years:
Structure. 2006 Feb;14(2):357-66.
Structural basis for sulfur relay to RNA mediated by heterohexameric TusBCD complex
Numata T, Fukai S, Ikeuchi Y, Suzuki T, Nureki O
Abstract
Uridine at wobble position 34 of tRNA(Lys), tRNA(Glu), and tRNA(Gln)
is exclusively modified into 2-thiouridine (s2U), which is crucial for
both precise codon recognition and recognition by the cognate
aminoacyl-tRNA synthetases. Recent Escherichia coli genetic studies
revealed that the products of five novel genes, tusABCDE, function in
the s2U modification. Here, we solved the 2.15 angstroms crystal
structure of the E. coli TusBCD complex, a sulfur transfer mediator,
forming a heterohexamer composed of a dimer of the
heterotrimer. Structure-based sequence alignment suggested two
putative active site Cys residues, Cys79 (in TusC) and Cys78 (in
TusD), which are exposed on the hexameric complex. In vivo mutant
analyses revealed that only Cys78, in the TusD subunit, participates
in sulfur transfer during the s2U modification process. Since the
single Cys acts as a catalytic residue, we proposed that TusBCD
mediates sulfur relay via a putative persulfide state of the TusD
subunit.
and
Mol Cell. 2006 Jan 6;21(1):97-108
Mechanistic insights into sulfur relay by multiple sulfur mediators
involved in thiouridine biosynthesis at tRNA wobble positions.
Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T.
Abstract
The wobble bases of bacterial tRNAs responsible for NNR codons are
modified to 5-methylaminomethyl-2-thiouridine (mnm5s2U). 2-thio
modification of mnm5s2U is required for accurate decoding and
essential for normal cell growth. We identified five genes yhhP, yheL,
yheM, yheN, and yccK (named tusA, tusB, tusC, tusD, and tusE,
respectively) that are essential for 2-thiouridylation of mnm5s2U by a
systematic genome-wide screen ("ribonucleome analysis"). Efficient
2-thiouridine formation in vitro was reconstituted with recombinant
TusA, a TusBCD complex, TusE, and previously identified IscS and
MnmA. The desulfurase activity of IscS is stimulated by TusA
binding. IscS transfers the persulfide sulfur to TusA. TusE binds
TusBCD complex and stimulates sulfur transfer from TusA to TusD. TusE
also interacts with an MnmA-tRNA complex. This study revealed that
2-thiouridine formation proceeds through a complex sulfur-relay system
composed of multiple sulfur mediators that select and facilitate
specific sulfur flow to 2-thiouridine from various pathways of sulfur
trafficking.
I would like to focus on fig|83333.1.peg.3280, which is currently assigned the
function "predicted DNA-binding transcriptional regulator" by some groups and
"Putative DNA-binding protein" within the SEED. If one looks at PFAM hits, you
would see
YheO-like PAS domain:
This family contains various hypothetical bacterial proteins that are
similar to the E. coli protein YheO. Their function is unknown, but
are likely to be involved in signalling based on the presence of this
PAS domain.
Here is a plot of the expression values for peg.80 versus peg.77, the last gene
in the putative operon.
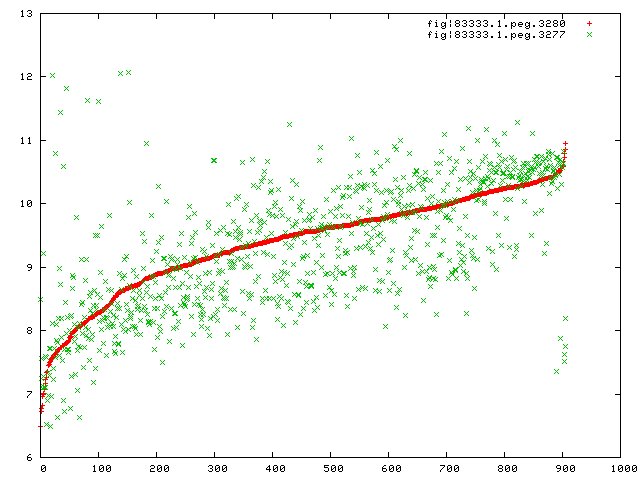
I am not sure if the gene is a regulatory gene, but it is closely related to the TusBCD
complex and its role of sulfur to to 2-thiouridine.
A Component of a Complex: E.coli Atomic Regulon 109 [ON=125 OFF=755]
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.279 |
Hypothetical protein YagQ |
|
| fig|83333.1.peg.280 |
Periplasmic aromatic aldehyde oxidoreductase, molybdenum binding subunit YagR |
CBSS-266117.6.peg.2476,Purine_Utilization,Putative_diaminopropionate_ammonia-lyase_cluster |
| fig|83333.1.peg.281 |
Periplasmic aromatic aldehyde oxidoreductase, FAD binding subunit YagS |
Purine_Utilization |
| fig|83333.1.peg.282 |
Periplasmic aromatic aldehyde oxidoreductase, iron-sulfur subunit YagT |
CBSS-266117.6.peg.2476,Purine_Utilization,Putative_diaminopropionate_ammonia-lyase_cluster |
If you search for domains that hit the hypothetical protein (YagQ), you will find
several that indicate the following:
xanthine dehydrogenase accessory protein XdhC
Members of this protein family are the accessory protein XdhC for insertion of the
molybdenum cofactor into the xanthine dehydrogenase large chain, XdhB, in bacteria.
This protein is not part of the mature xanthine dehydrogenase.
Xanthine dehydrogenase is an enzyme for purine catabolism,
from other purines to xanthine to urate to further breakdown products.
So, one would conjecture that this is a complex similar to xdhABCD, and YagQ plays a role analogous to XdhC.
The Glycine Cleavage System in Staph. aureus: Atomic Regulon 32 [ON=276 OFF=550]
The next example is from Staphylococcus aureus. It relates to a cluster that encodes the genes
related to a glycine catabolism complex (I believe).
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.323 |
Flavin-utilizing monoxygenases (EC 1.14.-.-) |
|
| fig|158878.1.peg.324 |
Glycine cleavage system H protein |
Glycine_and_Serine_Utilization,Glycine_cleavage_system,Photorespiration_(oxidative_C2_cycle) |
| fig|158878.1.peg.325 |
hypothetical protein |
|
| fig|158878.1.peg.326 |
NAD-dependent protein deacetylases, SIR2 family |
|
| fig|158878.1.peg.327 |
Lipoate-protein ligase A |
Dehydrogenase_complexes,Glycine_cleavage_system,Lipoic_acid_metabolism |
Here is a description of glycine cleavage H-proteins:
This is a family of glycine cleavage H-proteins, part of the glycine cleavage multienzyme complex (GCV) found in bacteria and the mitochondria of eukaryotes. GCV catalyses the catabolism of glycine in eukaryotes. A lipoyl group is attached to a completely conserved lysine residue. The H protein shuttles the methylamine group of glycine from the P protein to the T protein.
TIGR00545:lipoyltransferase and lipoate-protein ligase
One member of this group of proteins is bovine lipoyltransferase, which transfers the lipoyl group from lipoyl-AMP to the specific Lys of lipoate-dependent enzymes. However, it does not first activate lipoic acid with ATP to create lipoyl-AMP and pyrophosphate. Another member of this group, lipoate-protein ligase A from E. coli, catalyzes both the activation and the transfer of lipoate. Homology between the two is full-length, except for the bovine mitochondrial targeting signal, but is strongest toward the N-terminus.
Now let me quote from the abstract of a paper written in 1990:
JOURNAL OF BACTERIOLOGY, Oct. 1990, p. 6142-6144 Vol. 172, No. 10
The lpd Gene Product Functions as the L Protein in the
Escherichia coli Glycine Cleavage Enzyme System
PAULA S. STEIERT, LORRAINE T. STAUFFER, AND GEORGE V. STAUFFER
The enzyme serine hydroxymethyltransferase catalyzes the
conversion of serine to glycine and
5,10-methylenetetrahydrofolate and provides the major source of
one-carbon units. The oxidative cleavage of glycine to form
NH3, C02, and 5,10-methylenetetrahydrofolate provides a secondary
pathway for the biosynthesis of one-carbon units in mammalian and
bacterial systems. The glycine cleavage
(GCV) enzyme system has been described in Peptococcus
glycinophilus and consists of four protein components
designated P1, P2, P3, and P4. A GCV enzyme system that
can be induced by exogenous glycine has also been demonstrated
in Escherichia coli (9) and is located at minute 63 on
the linkage map (2). Although the system has not been well
characterized, at least one protein from E. coli interacts
catalytically with the P1 protein from P. glycinophilus in the
exchange of bicarbonate with 14C-labeled glycine, indicating
that the two systems may be similar (D. K. Ransom and
R. D. Sagers, Abstr. Annu. Meet. Am. Soc. Microbiol.
1974, P266, p. 189).
If one looks closely, it is clear that this is not the normal version of a glysine cleavage system.
The normal one is represented in other genes ()
Sveta Gerdes writes in her subsystem on glycine cleavage system:
"Gene "out of context":
In Chlamydia, Chlamidophyla, Streptococci, Bifidobacteria, Mycoplasma,
Acinetobacter, etc - a very close homolog of a Glycine cleavage
system H protein is present, often - in close proximity of
Lipoate-protein ligase A. However, other Glycine cleavage
system subunits are absent, the function of these proteins is
unknown"
We can pursue this issue by looking at the expression correlations between the four distinct
PEGs assigned the function Lipoate-protein ligase A and the two distinct PEGs labeled as
Glycine cleavage system H protein. If you do that, you find just weaker correlations (but no anti-correlations).
I believe that getting at the truth will require detailed examination of experimental conditions
to see which conditions distinguish expression of the genes.
I probably should not have included that example, but I think that it does illustrate
the style of reasoning that is required on every one of these atomic regulons. We will need to filter
and prioritize them, and each one requires a fair amount of study.
An Example Relating to Motility: Staph. aureus Atomic Regulon 2 [ON=229 OFF=12]
Pearson Coefficients:
| PEG |
peg.122 |
peg.1381 |
peg.1382 |
peg.2890 |
peg.2891 |
peg.2892 |
peg.2893 |
peg.2894 |
peg.2895 |
peg.2896 |
peg.2897 |
peg.2898 |
peg.2899 |
peg.2900 |
peg.2904 |
peg.2905 |
peg.2906 |
peg.2907 |
peg.2908 |
peg.2909 |
peg.2910 |
peg.2911 |
peg.2912 |
peg.2913 |
peg.2914 |
peg.2915 |
peg.2916 |
peg.2917 |
peg.2919 |
peg.2920 |
peg.2921 |
peg.2922 |
peg.2923 |
peg.2924 |
peg.2925 |
peg.2926 |
peg.2927 |
peg.2928 |
peg.2929 |
peg.2930 |
peg.2931 |
peg.2932 |
peg.2933 |
peg.2934 |
peg.2935 |
peg.2939 |
peg.2940 |
| fig|211586.9.peg.122 |
|
0.724 |
0.646 |
0.616 |
0.586 |
0.640 |
0.625 |
0.519 |
0.637 |
0.600 |
0.628 |
0.636 |
0.566 |
0.579 |
0.577 |
0.632 |
0.615 |
0.529 |
0.438 |
0.461 |
0.505 |
0.435 |
0.631 |
0.598 |
0.377 |
0.457 |
0.473 |
0.507 |
0.534 |
0.686 |
0.659 |
0.664 |
0.599 |
0.418 |
0.418 |
0.432 |
0.424 |
0.329 |
0.357 |
0.235 |
0.325 |
0.376 |
0.451 |
0.383 |
0.402 |
0.707 |
0.788 |
| fig|211586.9.peg.1381 |
0.724 |
|
0.778 |
0.631 |
0.507 |
0.536 |
0.536 |
0.507 |
0.698 |
0.718 |
0.737 |
0.637 |
0.520 |
0.532 |
0.413 |
0.590 |
0.645 |
0.501 |
0.491 |
0.483 |
0.273 |
0.151 |
0.533 |
0.615 |
0.377 |
0.494 |
0.371 |
0.377 |
0.673 |
0.740 |
0.702 |
0.689 |
0.633 |
0.522 |
0.504 |
0.430 |
0.494 |
0.460 |
0.424 |
0.403 |
0.439 |
0.513 |
0.565 |
0.561 |
0.531 |
0.824 |
0.810 |
| fig|211586.9.peg.1382 |
0.646 |
0.778 |
|
0.483 |
0.390 |
0.472 |
0.366 |
0.273 |
0.409 |
0.427 |
0.504 |
0.375 |
0.324 |
0.464 |
0.403 |
0.467 |
0.492 |
0.388 |
0.438 |
0.366 |
0.301 |
0.212 |
0.413 |
0.324 |
0.277 |
0.299 |
0.296 |
0.232 |
0.325 |
0.409 |
0.405 |
0.430 |
0.290 |
0.227 |
0.365 |
0.319 |
0.362 |
0.278 |
0.210 |
0.203 |
0.297 |
0.257 |
0.331 |
0.315 |
0.329 |
0.546 |
0.590 |
| fig|211586.9.peg.2890 |
0.616 |
0.631 |
0.483 |
|
0.868 |
0.835 |
0.808 |
0.695 |
0.795 |
0.759 |
0.764 |
0.759 |
0.586 |
0.424 |
0.674 |
0.747 |
0.627 |
0.640 |
0.546 |
0.540 |
0.588 |
0.316 |
0.664 |
0.704 |
0.365 |
0.457 |
0.504 |
0.476 |
0.687 |
0.709 |
0.683 |
0.660 |
0.553 |
0.541 |
0.650 |
0.601 |
0.448 |
0.431 |
0.361 |
0.305 |
0.325 |
0.538 |
0.557 |
0.524 |
0.483 |
0.540 |
0.566 |
| fig|211586.9.peg.2891 |
0.586 |
0.507 |
0.390 |
0.868 |
|
0.895 |
0.879 |
0.682 |
0.725 |
0.671 |
0.678 |
0.760 |
0.613 |
0.480 |
0.823 |
0.741 |
0.608 |
0.680 |
0.535 |
0.592 |
0.641 |
0.401 |
0.668 |
0.699 |
0.374 |
0.486 |
0.498 |
0.548 |
0.586 |
0.659 |
0.590 |
0.582 |
0.511 |
0.581 |
0.661 |
0.659 |
0.511 |
0.400 |
0.366 |
0.271 |
0.322 |
0.586 |
0.568 |
0.542 |
0.533 |
0.486 |
0.526 |
| fig|211586.9.peg.2892 |
0.640 |
0.536 |
0.472 |
0.835 |
0.895 |
|
0.885 |
0.736 |
0.747 |
0.669 |
0.715 |
0.767 |
0.674 |
0.578 |
0.795 |
0.782 |
0.682 |
0.734 |
0.677 |
0.669 |
0.725 |
0.535 |
0.792 |
0.719 |
0.456 |
0.516 |
0.592 |
0.631 |
0.563 |
0.610 |
0.601 |
0.609 |
0.476 |
0.553 |
0.683 |
0.707 |
0.593 |
0.481 |
0.401 |
0.309 |
0.345 |
0.572 |
0.563 |
0.506 |
0.529 |
0.503 |
0.555 |
| fig|211586.9.peg.2893 |
0.625 |
0.536 |
0.366 |
0.808 |
0.879 |
0.885 |
|
0.819 |
0.798 |
0.665 |
0.698 |
0.817 |
0.749 |
0.592 |
0.749 |
0.765 |
0.731 |
0.792 |
0.573 |
0.664 |
0.656 |
0.575 |
0.773 |
0.784 |
0.510 |
0.598 |
0.633 |
0.680 |
0.592 |
0.751 |
0.647 |
0.614 |
0.612 |
0.697 |
0.691 |
0.722 |
0.604 |
0.494 |
0.541 |
0.415 |
0.445 |
0.682 |
0.692 |
0.623 |
0.636 |
0.515 |
0.577 |
| fig|211586.9.peg.2894 |
0.519 |
0.507 |
0.273 |
0.695 |
0.682 |
0.736 |
0.819 |
|
0.856 |
0.776 |
0.792 |
0.842 |
0.814 |
0.595 |
0.505 |
0.663 |
0.755 |
0.804 |
0.689 |
0.716 |
0.545 |
0.543 |
0.835 |
0.874 |
0.610 |
0.620 |
0.676 |
0.698 |
0.761 |
0.778 |
0.789 |
0.752 |
0.694 |
0.753 |
0.717 |
0.761 |
0.636 |
0.672 |
0.713 |
0.611 |
0.548 |
0.720 |
0.736 |
0.650 |
0.659 |
0.576 |
0.604 |
| fig|211586.9.peg.2895 |
0.637 |
0.698 |
0.409 |
0.795 |
0.725 |
0.747 |
0.798 |
0.856 |
|
0.908 |
0.897 |
0.865 |
0.744 |
0.529 |
0.509 |
0.675 |
0.703 |
0.705 |
0.606 |
0.582 |
0.448 |
0.327 |
0.769 |
0.862 |
0.500 |
0.583 |
0.568 |
0.598 |
0.823 |
0.869 |
0.846 |
0.790 |
0.739 |
0.726 |
0.667 |
0.641 |
0.539 |
0.566 |
0.577 |
0.518 |
0.466 |
0.668 |
0.711 |
0.653 |
0.612 |
0.702 |
0.716 |
| fig|211586.9.peg.2896 |
0.600 |
0.718 |
0.427 |
0.759 |
0.671 |
0.669 |
0.665 |
0.776 |
0.908 |
|
0.946 |
0.814 |
0.668 |
0.468 |
0.470 |
0.612 |
0.619 |
0.609 |
0.586 |
0.519 |
0.376 |
0.216 |
0.696 |
0.808 |
0.387 |
0.536 |
0.498 |
0.483 |
0.843 |
0.786 |
0.793 |
0.769 |
0.657 |
0.642 |
0.681 |
0.611 |
0.495 |
0.550 |
0.518 |
0.532 |
0.431 |
0.603 |
0.629 |
0.602 |
0.555 |
0.719 |
0.711 |
| fig|211586.9.peg.2897 |
0.628 |
0.737 |
0.504 |
0.764 |
0.678 |
0.715 |
0.698 |
0.792 |
0.897 |
0.946 |
|
0.820 |
0.705 |
0.577 |
0.537 |
0.612 |
0.635 |
0.668 |
0.633 |
0.591 |
0.429 |
0.306 |
0.754 |
0.833 |
0.461 |
0.588 |
0.567 |
0.558 |
0.803 |
0.790 |
0.766 |
0.766 |
0.645 |
0.643 |
0.708 |
0.669 |
0.538 |
0.563 |
0.580 |
0.547 |
0.471 |
0.593 |
0.641 |
0.607 |
0.605 |
0.731 |
0.744 |
| fig|211586.9.peg.2898 |
0.636 |
0.637 |
0.375 |
0.759 |
0.760 |
0.767 |
0.817 |
0.842 |
0.865 |
0.814 |
0.820 |
|
0.846 |
0.684 |
0.615 |
0.807 |
0.817 |
0.818 |
0.734 |
0.745 |
0.624 |
0.452 |
0.809 |
0.896 |
0.631 |
0.724 |
0.697 |
0.743 |
0.821 |
0.827 |
0.807 |
0.779 |
0.720 |
0.790 |
0.765 |
0.760 |
0.763 |
0.718 |
0.693 |
0.597 |
0.614 |
0.771 |
0.791 |
0.746 |
0.709 |
0.690 |
0.717 |
| fig|211586.9.peg.2899 |
0.566 |
0.520 |
0.324 |
0.586 |
0.613 |
0.674 |
0.749 |
0.814 |
0.744 |
0.668 |
0.705 |
0.846 |
|
0.814 |
0.510 |
0.702 |
0.802 |
0.855 |
0.786 |
0.857 |
0.622 |
0.624 |
0.869 |
0.866 |
0.825 |
0.848 |
0.846 |
0.895 |
0.677 |
0.741 |
0.713 |
0.699 |
0.600 |
0.761 |
0.756 |
0.814 |
0.804 |
0.765 |
0.806 |
0.744 |
0.799 |
0.722 |
0.806 |
0.714 |
0.777 |
0.613 |
0.678 |
| fig|211586.9.peg.2900 |
0.579 |
0.532 |
0.464 |
0.424 |
0.480 |
0.578 |
0.592 |
0.595 |
0.529 |
0.468 |
0.577 |
0.684 |
0.814 |
|
0.529 |
0.613 |
0.727 |
0.714 |
0.695 |
0.803 |
0.606 |
0.654 |
0.773 |
0.708 |
0.785 |
0.837 |
0.753 |
0.832 |
0.472 |
0.589 |
0.513 |
0.535 |
0.439 |
0.594 |
0.630 |
0.701 |
0.820 |
0.626 |
0.729 |
0.648 |
0.744 |
0.545 |
0.659 |
0.596 |
0.698 |
0.575 |
0.661 |
| fig|211586.9.peg.2904 |
0.577 |
0.413 |
0.403 |
0.674 |
0.823 |
0.795 |
0.749 |
0.505 |
0.509 |
0.470 |
0.537 |
0.615 |
0.510 |
0.529 |
|
0.681 |
0.545 |
0.658 |
0.531 |
0.559 |
0.719 |
0.466 |
0.609 |
0.567 |
0.318 |
0.436 |
0.491 |
0.534 |
0.381 |
0.469 |
0.402 |
0.447 |
0.338 |
0.404 |
0.597 |
0.625 |
0.495 |
0.320 |
0.314 |
0.200 |
0.275 |
0.472 |
0.465 |
0.429 |
0.475 |
0.392 |
0.456 |
| fig|211586.9.peg.2905 |
0.632 |
0.590 |
0.467 |
0.747 |
0.741 |
0.782 |
0.765 |
0.663 |
0.675 |
0.612 |
0.612 |
0.807 |
0.702 |
0.613 |
0.681 |
|
0.856 |
0.771 |
0.707 |
0.683 |
0.733 |
0.521 |
0.706 |
0.715 |
0.559 |
0.578 |
0.678 |
0.641 |
0.618 |
0.639 |
0.635 |
0.590 |
0.518 |
0.591 |
0.656 |
0.668 |
0.687 |
0.627 |
0.525 |
0.417 |
0.518 |
0.640 |
0.663 |
0.602 |
0.541 |
0.525 |
0.579 |
| fig|211586.9.peg.2906 |
0.615 |
0.645 |
0.492 |
0.627 |
0.608 |
0.682 |
0.731 |
0.755 |
0.703 |
0.619 |
0.635 |
0.817 |
0.802 |
0.727 |
0.545 |
0.856 |
|
0.806 |
0.745 |
0.752 |
0.649 |
0.571 |
0.755 |
0.765 |
0.693 |
0.690 |
0.708 |
0.709 |
0.665 |
0.722 |
0.705 |
0.659 |
0.598 |
0.720 |
0.706 |
0.700 |
0.767 |
0.747 |
0.693 |
0.595 |
0.680 |
0.716 |
0.767 |
0.686 |
0.671 |
0.615 |
0.670 |
| fig|211586.9.peg.2907 |
0.529 |
0.501 |
0.388 |
0.640 |
0.680 |
0.734 |
0.792 |
0.804 |
0.705 |
0.609 |
0.668 |
0.818 |
0.855 |
0.714 |
0.658 |
0.771 |
0.806 |
|
0.825 |
0.815 |
0.714 |
0.663 |
0.839 |
0.802 |
0.706 |
0.692 |
0.796 |
0.771 |
0.606 |
0.662 |
0.647 |
0.645 |
0.528 |
0.679 |
0.765 |
0.842 |
0.731 |
0.718 |
0.737 |
0.614 |
0.642 |
0.721 |
0.757 |
0.655 |
0.702 |
0.542 |
0.595 |
| fig|211586.9.peg.2908 |
0.438 |
0.491 |
0.438 |
0.546 |
0.535 |
0.677 |
0.573 |
0.689 |
0.606 |
0.586 |
0.633 |
0.734 |
0.786 |
0.695 |
0.531 |
0.707 |
0.745 |
0.825 |
|
0.816 |
0.675 |
0.526 |
0.794 |
0.710 |
0.694 |
0.647 |
0.754 |
0.733 |
0.580 |
0.482 |
0.587 |
0.613 |
0.386 |
0.518 |
0.709 |
0.776 |
0.758 |
0.753 |
0.633 |
0.621 |
0.623 |
0.606 |
0.652 |
0.587 |
0.616 |
0.485 |
0.523 |
| fig|211586.9.peg.2909 |
0.461 |
0.483 |
0.366 |
0.540 |
0.592 |
0.669 |
0.664 |
0.716 |
0.582 |
0.519 |
0.591 |
0.745 |
0.857 |
0.803 |
0.559 |
0.683 |
0.752 |
0.815 |
0.816 |
|
0.648 |
0.558 |
0.796 |
0.768 |
0.833 |
0.832 |
0.776 |
0.872 |
0.626 |
0.636 |
0.658 |
0.684 |
0.548 |
0.752 |
0.806 |
0.869 |
0.867 |
0.797 |
0.775 |
0.689 |
0.770 |
0.779 |
0.813 |
0.742 |
0.831 |
0.592 |
0.633 |
| fig|211586.9.peg.2910 |
0.505 |
0.273 |
0.301 |
0.588 |
0.641 |
0.725 |
0.656 |
0.545 |
0.448 |
0.376 |
0.429 |
0.624 |
0.622 |
0.606 |
0.719 |
0.733 |
0.649 |
0.714 |
0.675 |
0.648 |
|
0.722 |
0.718 |
0.575 |
0.527 |
0.467 |
0.659 |
0.645 |
0.369 |
0.405 |
0.435 |
0.446 |
0.275 |
0.357 |
0.542 |
0.658 |
0.594 |
0.484 |
0.395 |
0.254 |
0.366 |
0.395 |
0.409 |
0.328 |
0.379 |
0.326 |
0.423 |
| fig|211586.9.peg.2911 |
0.435 |
0.151 |
0.212 |
0.316 |
0.401 |
0.535 |
0.575 |
0.543 |
0.327 |
0.216 |
0.306 |
0.452 |
0.624 |
0.654 |
0.466 |
0.521 |
0.571 |
0.663 |
0.526 |
0.558 |
0.722 |
|
0.709 |
0.486 |
0.594 |
0.469 |
0.700 |
0.635 |
0.157 |
0.306 |
0.264 |
0.263 |
0.156 |
0.311 |
0.401 |
0.609 |
0.525 |
0.435 |
0.507 |
0.360 |
0.421 |
0.290 |
0.343 |
0.209 |
0.319 |
0.193 |
0.334 |
| fig|211586.9.peg.2912 |
0.631 |
0.533 |
0.413 |
0.664 |
0.668 |
0.792 |
0.773 |
0.835 |
0.769 |
0.696 |
0.754 |
0.809 |
0.869 |
0.773 |
0.609 |
0.706 |
0.755 |
0.839 |
0.794 |
0.796 |
0.718 |
0.709 |
|
0.875 |
0.711 |
0.700 |
0.807 |
0.817 |
0.632 |
0.668 |
0.694 |
0.691 |
0.510 |
0.618 |
0.710 |
0.816 |
0.719 |
0.669 |
0.678 |
0.591 |
0.580 |
0.604 |
0.661 |
0.558 |
0.621 |
0.583 |
0.652 |
| fig|211586.9.peg.2913 |
0.598 |
0.615 |
0.324 |
0.704 |
0.699 |
0.719 |
0.784 |
0.874 |
0.862 |
0.808 |
0.833 |
0.896 |
0.866 |
0.708 |
0.567 |
0.715 |
0.765 |
0.802 |
0.710 |
0.768 |
0.575 |
0.486 |
0.875 |
|
0.696 |
0.744 |
0.748 |
0.791 |
0.794 |
0.821 |
0.794 |
0.758 |
0.699 |
0.768 |
0.749 |
0.796 |
0.734 |
0.725 |
0.763 |
0.682 |
0.629 |
0.754 |
0.796 |
0.734 |
0.722 |
0.675 |
0.694 |
| fig|211586.9.peg.2914 |
0.377 |
0.377 |
0.277 |
0.365 |
0.374 |
0.456 |
0.510 |
0.610 |
0.500 |
0.387 |
0.461 |
0.631 |
0.825 |
0.785 |
0.318 |
0.559 |
0.693 |
0.706 |
0.694 |
0.833 |
0.527 |
0.594 |
0.711 |
0.696 |
|
0.849 |
0.842 |
0.873 |
0.532 |
0.556 |
0.563 |
0.567 |
0.456 |
0.656 |
0.656 |
0.754 |
0.808 |
0.815 |
0.816 |
0.741 |
0.858 |
0.645 |
0.724 |
0.608 |
0.672 |
0.471 |
0.534 |
| fig|211586.9.peg.2915 |
0.457 |
0.494 |
0.299 |
0.457 |
0.486 |
0.516 |
0.598 |
0.620 |
0.583 |
0.536 |
0.588 |
0.724 |
0.848 |
0.837 |
0.436 |
0.578 |
0.690 |
0.692 |
0.647 |
0.832 |
0.467 |
0.469 |
0.700 |
0.744 |
0.849 |
|
0.780 |
0.867 |
0.634 |
0.686 |
0.604 |
0.616 |
0.567 |
0.795 |
0.782 |
0.774 |
0.861 |
0.771 |
0.843 |
0.814 |
0.889 |
0.744 |
0.821 |
0.785 |
0.853 |
0.618 |
0.647 |
| fig|211586.9.peg.2916 |
0.473 |
0.371 |
0.296 |
0.504 |
0.498 |
0.592 |
0.633 |
0.676 |
0.568 |
0.498 |
0.567 |
0.697 |
0.846 |
0.753 |
0.491 |
0.678 |
0.708 |
0.796 |
0.754 |
0.776 |
0.659 |
0.700 |
0.807 |
0.748 |
0.842 |
0.780 |
|
0.878 |
0.537 |
0.546 |
0.540 |
0.526 |
0.408 |
0.566 |
0.690 |
0.804 |
0.745 |
0.752 |
0.776 |
0.730 |
0.742 |
0.596 |
0.691 |
0.590 |
0.620 |
0.418 |
0.493 |
| fig|211586.9.peg.2917 |
0.507 |
0.377 |
0.232 |
0.476 |
0.548 |
0.631 |
0.680 |
0.698 |
0.598 |
0.483 |
0.558 |
0.743 |
0.895 |
0.832 |
0.534 |
0.641 |
0.709 |
0.771 |
0.733 |
0.872 |
0.645 |
0.635 |
0.817 |
0.791 |
0.873 |
0.867 |
0.878 |
|
0.572 |
0.629 |
0.599 |
0.607 |
0.524 |
0.694 |
0.717 |
0.818 |
0.814 |
0.733 |
0.788 |
0.685 |
0.765 |
0.678 |
0.743 |
0.657 |
0.728 |
0.511 |
0.579 |
| fig|211586.9.peg.2919 |
0.534 |
0.673 |
0.325 |
0.687 |
0.586 |
0.563 |
0.592 |
0.761 |
0.823 |
0.843 |
0.803 |
0.821 |
0.677 |
0.472 |
0.381 |
0.618 |
0.665 |
0.606 |
0.580 |
0.626 |
0.369 |
0.157 |
0.632 |
0.794 |
0.532 |
0.634 |
0.537 |
0.572 |
|
0.869 |
0.904 |
0.872 |
0.811 |
0.786 |
0.726 |
0.639 |
0.599 |
0.707 |
0.650 |
0.615 |
0.572 |
0.744 |
0.748 |
0.733 |
0.671 |
0.758 |
0.723 |
| fig|211586.9.peg.2920 |
0.686 |
0.740 |
0.409 |
0.709 |
0.659 |
0.610 |
0.751 |
0.778 |
0.869 |
0.786 |
0.790 |
0.827 |
0.741 |
0.589 |
0.469 |
0.639 |
0.722 |
0.662 |
0.482 |
0.636 |
0.405 |
0.306 |
0.668 |
0.821 |
0.556 |
0.686 |
0.546 |
0.629 |
0.869 |
|
0.908 |
0.858 |
0.908 |
0.839 |
0.659 |
0.616 |
0.582 |
0.580 |
0.658 |
0.555 |
0.582 |
0.736 |
0.793 |
0.760 |
0.742 |
0.824 |
0.821 |
| fig|211586.9.peg.2921 |
0.659 |
0.702 |
0.405 |
0.683 |
0.590 |
0.601 |
0.647 |
0.789 |
0.846 |
0.793 |
0.766 |
0.807 |
0.713 |
0.513 |
0.402 |
0.635 |
0.705 |
0.647 |
0.587 |
0.658 |
0.435 |
0.264 |
0.694 |
0.794 |
0.563 |
0.604 |
0.540 |
0.599 |
0.904 |
0.908 |
|
0.964 |
0.895 |
0.756 |
0.613 |
0.585 |
0.570 |
0.642 |
0.600 |
0.522 |
0.535 |
0.684 |
0.726 |
0.671 |
0.653 |
0.841 |
0.827 |
| fig|211586.9.peg.2922 |
0.664 |
0.689 |
0.430 |
0.660 |
0.582 |
0.609 |
0.614 |
0.752 |
0.790 |
0.769 |
0.766 |
0.779 |
0.699 |
0.535 |
0.447 |
0.590 |
0.659 |
0.645 |
0.613 |
0.684 |
0.446 |
0.263 |
0.691 |
0.758 |
0.567 |
0.616 |
0.526 |
0.607 |
0.872 |
0.858 |
0.964 |
|
0.862 |
0.715 |
0.637 |
0.608 |
0.569 |
0.642 |
0.583 |
0.500 |
0.531 |
0.651 |
0.686 |
0.623 |
0.654 |
0.874 |
0.856 |
| fig|211586.9.peg.2923 |
0.599 |
0.633 |
0.290 |
0.553 |
0.511 |
0.476 |
0.612 |
0.694 |
0.739 |
0.657 |
0.645 |
0.720 |
0.600 |
0.439 |
0.338 |
0.518 |
0.598 |
0.528 |
0.386 |
0.548 |
0.275 |
0.156 |
0.510 |
0.699 |
0.456 |
0.567 |
0.408 |
0.524 |
0.811 |
0.908 |
0.895 |
0.862 |
|
0.796 |
0.511 |
0.464 |
0.495 |
0.519 |
0.567 |
0.453 |
0.472 |
0.695 |
0.718 |
0.692 |
0.661 |
0.815 |
0.770 |
| fig|211586.9.peg.2924 |
0.418 |
0.522 |
0.227 |
0.541 |
0.581 |
0.553 |
0.697 |
0.753 |
0.726 |
0.642 |
0.643 |
0.790 |
0.761 |
0.594 |
0.404 |
0.591 |
0.720 |
0.679 |
0.518 |
0.752 |
0.357 |
0.311 |
0.618 |
0.768 |
0.656 |
0.795 |
0.566 |
0.694 |
0.786 |
0.839 |
0.756 |
0.715 |
0.796 |
|
0.796 |
0.716 |
0.752 |
0.742 |
0.796 |
0.714 |
0.743 |
0.914 |
0.905 |
0.878 |
0.876 |
0.659 |
0.650 |
| fig|211586.9.peg.2925 |
0.418 |
0.504 |
0.365 |
0.650 |
0.661 |
0.683 |
0.691 |
0.717 |
0.667 |
0.681 |
0.708 |
0.765 |
0.756 |
0.630 |
0.597 |
0.656 |
0.706 |
0.765 |
0.709 |
0.806 |
0.542 |
0.401 |
0.710 |
0.749 |
0.656 |
0.782 |
0.690 |
0.717 |
0.726 |
0.659 |
0.613 |
0.637 |
0.511 |
0.796 |
|
0.897 |
0.775 |
0.812 |
0.772 |
0.747 |
0.729 |
0.847 |
0.832 |
0.802 |
0.819 |
0.548 |
0.566 |
| fig|211586.9.peg.2926 |
0.432 |
0.430 |
0.319 |
0.601 |
0.659 |
0.707 |
0.722 |
0.761 |
0.641 |
0.611 |
0.669 |
0.760 |
0.814 |
0.701 |
0.625 |
0.668 |
0.700 |
0.842 |
0.776 |
0.869 |
0.658 |
0.609 |
0.816 |
0.796 |
0.754 |
0.774 |
0.804 |
0.818 |
0.639 |
0.616 |
0.585 |
0.608 |
0.464 |
0.716 |
0.897 |
|
0.818 |
0.813 |
0.808 |
0.711 |
0.701 |
0.790 |
0.784 |
0.705 |
0.751 |
0.496 |
0.542 |
| fig|211586.9.peg.2927 |
0.424 |
0.494 |
0.362 |
0.448 |
0.511 |
0.593 |
0.604 |
0.636 |
0.539 |
0.495 |
0.538 |
0.763 |
0.804 |
0.820 |
0.495 |
0.687 |
0.767 |
0.731 |
0.758 |
0.867 |
0.594 |
0.525 |
0.719 |
0.734 |
0.808 |
0.861 |
0.745 |
0.814 |
0.599 |
0.582 |
0.570 |
0.569 |
0.495 |
0.752 |
0.775 |
0.818 |
|
0.852 |
0.804 |
0.749 |
0.817 |
0.786 |
0.800 |
0.756 |
0.785 |
0.552 |
0.586 |
| fig|211586.9.peg.2928 |
0.329 |
0.460 |
0.278 |
0.431 |
0.400 |
0.481 |
0.494 |
0.672 |
0.566 |
0.550 |
0.563 |
0.718 |
0.765 |
0.626 |
0.320 |
0.627 |
0.747 |
0.718 |
0.753 |
0.797 |
0.484 |
0.435 |
0.669 |
0.725 |
0.815 |
0.771 |
0.752 |
0.733 |
0.707 |
0.580 |
0.642 |
0.642 |
0.519 |
0.742 |
0.812 |
0.813 |
0.852 |
|
0.862 |
0.823 |
0.831 |
0.809 |
0.806 |
0.733 |
0.724 |
0.543 |
0.545 |
| fig|211586.9.peg.2929 |
0.357 |
0.424 |
0.210 |
0.361 |
0.366 |
0.401 |
0.541 |
0.713 |
0.577 |
0.518 |
0.580 |
0.693 |
0.806 |
0.729 |
0.314 |
0.525 |
0.693 |
0.737 |
0.633 |
0.775 |
0.395 |
0.507 |
0.678 |
0.763 |
0.816 |
0.843 |
0.776 |
0.788 |
0.650 |
0.658 |
0.600 |
0.583 |
0.567 |
0.796 |
0.772 |
0.808 |
0.804 |
0.862 |
|
0.892 |
0.857 |
0.799 |
0.851 |
0.781 |
0.800 |
0.538 |
0.566 |
| fig|211586.9.peg.2930 |
0.235 |
0.403 |
0.203 |
0.305 |
0.271 |
0.309 |
0.415 |
0.611 |
0.518 |
0.532 |
0.547 |
0.597 |
0.744 |
0.648 |
0.200 |
0.417 |
0.595 |
0.614 |
0.621 |
0.689 |
0.254 |
0.360 |
0.591 |
0.682 |
0.741 |
0.814 |
0.730 |
0.685 |
0.615 |
0.555 |
0.522 |
0.500 |
0.453 |
0.714 |
0.747 |
0.711 |
0.749 |
0.823 |
0.892 |
|
0.879 |
0.732 |
0.814 |
0.786 |
0.791 |
0.465 |
0.469 |
| fig|211586.9.peg.2931 |
0.325 |
0.439 |
0.297 |
0.325 |
0.322 |
0.345 |
0.445 |
0.548 |
0.466 |
0.431 |
0.471 |
0.614 |
0.799 |
0.744 |
0.275 |
0.518 |
0.680 |
0.642 |
0.623 |
0.770 |
0.366 |
0.421 |
0.580 |
0.629 |
0.858 |
0.889 |
0.742 |
0.765 |
0.572 |
0.582 |
0.535 |
0.531 |
0.472 |
0.743 |
0.729 |
0.701 |
0.817 |
0.831 |
0.857 |
0.879 |
|
0.715 |
0.801 |
0.758 |
0.808 |
0.515 |
0.548 |
| fig|211586.9.peg.2932 |
0.376 |
0.513 |
0.257 |
0.538 |
0.586 |
0.572 |
0.682 |
0.720 |
0.668 |
0.603 |
0.593 |
0.771 |
0.722 |
0.545 |
0.472 |
0.640 |
0.716 |
0.721 |
0.606 |
0.779 |
0.395 |
0.290 |
0.604 |
0.754 |
0.645 |
0.744 |
0.596 |
0.678 |
0.744 |
0.736 |
0.684 |
0.651 |
0.695 |
0.914 |
0.847 |
0.790 |
0.786 |
0.809 |
0.799 |
0.732 |
0.715 |
|
0.939 |
0.912 |
0.864 |
0.577 |
0.558 |
| fig|211586.9.peg.2933 |
0.451 |
0.565 |
0.331 |
0.557 |
0.568 |
0.563 |
0.692 |
0.736 |
0.711 |
0.629 |
0.641 |
0.791 |
0.806 |
0.659 |
0.465 |
0.663 |
0.767 |
0.757 |
0.652 |
0.813 |
0.409 |
0.343 |
0.661 |
0.796 |
0.724 |
0.821 |
0.691 |
0.743 |
0.748 |
0.793 |
0.726 |
0.686 |
0.718 |
0.905 |
0.832 |
0.784 |
0.800 |
0.806 |
0.851 |
0.814 |
0.801 |
0.939 |
|
0.955 |
0.923 |
0.632 |
0.631 |
| fig|211586.9.peg.2934 |
0.383 |
0.561 |
0.315 |
0.524 |
0.542 |
0.506 |
0.623 |
0.650 |
0.653 |
0.602 |
0.607 |
0.746 |
0.714 |
0.596 |
0.429 |
0.602 |
0.686 |
0.655 |
0.587 |
0.742 |
0.328 |
0.209 |
0.558 |
0.734 |
0.608 |
0.785 |
0.590 |
0.657 |
0.733 |
0.760 |
0.671 |
0.623 |
0.692 |
0.878 |
0.802 |
0.705 |
0.756 |
0.733 |
0.781 |
0.786 |
0.758 |
0.912 |
0.955 |
|
0.935 |
0.602 |
0.575 |
| fig|211586.9.peg.2935 |
0.402 |
0.531 |
0.329 |
0.483 |
0.533 |
0.529 |
0.636 |
0.659 |
0.612 |
0.555 |
0.605 |
0.709 |
0.777 |
0.698 |
0.475 |
0.541 |
0.671 |
0.702 |
0.616 |
0.831 |
0.379 |
0.319 |
0.621 |
0.722 |
0.672 |
0.853 |
0.620 |
0.728 |
0.671 |
0.742 |
0.653 |
0.654 |
0.661 |
0.876 |
0.819 |
0.751 |
0.785 |
0.724 |
0.800 |
0.791 |
0.808 |
0.864 |
0.923 |
0.935 |
|
0.643 |
0.636 |
| fig|211586.9.peg.2939 |
0.707 |
0.824 |
0.546 |
0.540 |
0.486 |
0.503 |
0.515 |
0.576 |
0.702 |
0.719 |
0.731 |
0.690 |
0.613 |
0.575 |
0.392 |
0.525 |
0.615 |
0.542 |
0.485 |
0.592 |
0.326 |
0.193 |
0.583 |
0.675 |
0.471 |
0.618 |
0.418 |
0.511 |
0.758 |
0.824 |
0.841 |
0.874 |
0.815 |
0.659 |
0.548 |
0.496 |
0.552 |
0.543 |
0.538 |
0.465 |
0.515 |
0.577 |
0.632 |
0.602 |
0.643 |
|
0.958 |
| fig|211586.9.peg.2940 |
0.788 |
0.810 |
0.590 |
0.566 |
0.526 |
0.555 |
0.577 |
0.604 |
0.716 |
0.711 |
0.744 |
0.717 |
0.678 |
0.661 |
0.456 |
0.579 |
0.670 |
0.595 |
0.523 |
0.633 |
0.423 |
0.334 |
0.652 |
0.694 |
0.534 |
0.647 |
0.493 |
0.579 |
0.723 |
0.821 |
0.827 |
0.856 |
0.770 |
0.650 |
0.566 |
0.542 |
0.586 |
0.545 |
0.566 |
0.469 |
0.548 |
0.558 |
0.631 |
0.575 |
0.636 |
0.958 |
|
Functions in Shewanella oneidensis MR-1
| PEG |
Function |
Subsystems |
| fig|211586.9.peg.122 |
Flagellar biosynthesis protein FliL |
Flagellum |
| fig|211586.9.peg.1381 |
Flagellar motor rotation protein MotA |
Flagellum |
| fig|211586.9.peg.1382 |
Flagellar motor rotation protein MotB |
CBSS-323098.3.peg.2823,Flagellum |
| fig|211586.9.peg.2890 |
CheW domain protein |
|
| fig|211586.9.peg.2891 |
ParA family protein |
|
| fig|211586.9.peg.2892 |
hypothetical protein |
|
| fig|211586.9.peg.2893 |
Chemotaxis response regulator protein-glutamate methylesterase CheB (EC 3.1.1.61) |
|
| fig|211586.9.peg.2894 |
Signal transduction histidine kinase CheA (EC 2.7.3.-) |
|
| fig|211586.9.peg.2895 |
Chemotaxis response - phosphatase CheZ |
|
| fig|211586.9.peg.2896 |
Chemotaxis regulator - transmits chemoreceptor signals to flagelllar motor components CheY |
|
| fig|211586.9.peg.2897 |
RNA polymerase sigma factor for flagellar operon |
Flagellum,Transcription_initiation,_bacterial_sigma_factors |
| fig|211586.9.peg.2898 |
Flagellar synthesis regulator FleN |
Flagellum |
| fig|211586.9.peg.2899 |
Flagellar biosynthesis protein FlhF |
Flagellum |
| fig|211586.9.peg.2900 |
Flagellar biosynthesis protein FlhA |
Flagellum |
| fig|211586.9.peg.2904 |
Flagellar biosynthesis protein FliP |
Flagellum |
| fig|211586.9.peg.2905 |
Flagellar biosynthesis protein FliO |
Flagellum |
| fig|211586.9.peg.2906 |
Flagellar motor switch protein FliN |
Flagellum |
| fig|211586.9.peg.2907 |
Flagellar motor switch protein FliM |
Flagellum |
| fig|211586.9.peg.2908 |
Flagellar biosynthesis protein FliL |
Flagellum |
| fig|211586.9.peg.2909 |
Flagellar hook-length control protein FliK |
Flagellum |
| fig|211586.9.peg.2910 |
Flagellar protein FliJ |
Flagellum |
| fig|211586.9.peg.2911 |
Flagellum-specific ATP synthase FliI |
Flagellum |
| fig|211586.9.peg.2912 |
Flagellar assembly protein fliH |
|
| fig|211586.9.peg.2913 |
Flagellar motor switch protein fliG |
|
| fig|211586.9.peg.2914 |
Flagellar M-ring protein FliF |
Flagellum |
| fig|211586.9.peg.2915 |
Flagellar hook-basal body complex protein fliE |
|
| fig|211586.9.peg.2916 |
Flagellar regulatory protein FleQ |
Flagellum |
| fig|211586.9.peg.2917 |
Flagellar sensor histidine kinase FleS |
Flagellum |
| fig|211586.9.peg.2919 |
Flagellar biosynthesis protein FliS |
Flagellum |
| fig|211586.9.peg.2920 |
hypothetical protein |
|
| fig|211586.9.peg.2921 |
Flagellar hook-associated protein FliD |
Flagellum |
| fig|211586.9.peg.2922 |
Flagellin protein FlaG |
Flagellum |
| fig|211586.9.peg.2923 |
Flagellin protein FlaA |
Flagellum |
| fig|211586.9.peg.2924 |
Flagellin protein FlaA |
Flagellum |
| fig|211586.9.peg.2925 |
Flagellar hook-associated protein FlgL |
Flagellum |
| fig|211586.9.peg.2926 |
Flagellar hook-associated protein FlgK |
Flagellum |
| fig|211586.9.peg.2927 |
Flagellar protein FlgJ [peptidoglycan hydrolase] (EC 3.2.1.-) |
Flagellum |
| fig|211586.9.peg.2928 |
Flagellar P-ring protein FlgI |
Flagellum |
| fig|211586.9.peg.2929 |
Flagellar L-ring protein FlgH |
Flagellum |
| fig|211586.9.peg.2930 |
Flagellar basal-body rod protein FlgG |
Flagellum |
| fig|211586.9.peg.2931 |
Flagellar basal-body rod protein FlgF |
Flagellum |
| fig|211586.9.peg.2932 |
Flagellar hook protein FlgE |
Flagellum |
| fig|211586.9.peg.2933 |
Flagellar basal-body rod modification protein FlgD |
Flagellum |
| fig|211586.9.peg.2934 |
Flagellar basal-body rod protein flgC |
|
| fig|211586.9.peg.2935 |
Flagellar basal-body rod protein flgB |
|
| fig|211586.9.peg.2939 |
Negative regulator of flagellin synthesis FlgM |
Flagellum |
| fig|211586.9.peg.2940 |
Flagellar biosynthesis protein FlgN |
Flagellum |
Look at PEG 2892. It is clearly related to flagellar motility. However, it is worth reflecting that PEG 2891
is also in the same huge group and is now annotated as a ParA family protein (which is a protein that is
often described as relating to chromosome partitioning. When that annotation is made precise, it will probably
reflect a role in flagellar motility, as well.
Here is an example from Thermus thermophilus HB8:
Pegs in Atomic Regulon 4 [ON=76 OFF=37]
Pearson Coefficients:
| PEG |
peg.776 |
peg.777 |
peg.2037 |
peg.2038 |
peg.2039 |
peg.2040 |
peg.2041 |
peg.2042 |
peg.2043 |
peg.2044 |
peg.2045 |
peg.2046 |
peg.2047 |
peg.2048 |
peg.2049 |
| fig|300852.3.peg.776 |
|
0.940 |
0.721 |
0.845 |
0.882 |
0.863 |
0.808 |
0.814 |
0.878 |
0.849 |
0.765 |
0.824 |
0.800 |
0.824 |
0.712 |
| fig|300852.3.peg.777 |
0.940 |
|
0.629 |
0.821 |
0.860 |
0.845 |
0.843 |
0.860 |
0.861 |
0.854 |
0.827 |
0.869 |
0.865 |
0.882 |
0.821 |
| fig|300852.3.peg.2037 |
0.721 |
0.629 |
|
0.886 |
0.851 |
0.898 |
0.813 |
0.734 |
0.845 |
0.791 |
0.666 |
0.663 |
0.716 |
0.690 |
0.584 |
| fig|300852.3.peg.2038 |
0.845 |
0.821 |
0.886 |
|
0.962 |
0.963 |
0.940 |
0.924 |
0.945 |
0.939 |
0.873 |
0.890 |
0.864 |
0.875 |
0.829 |
| fig|300852.3.peg.2039 |
0.882 |
0.860 |
0.851 |
0.962 |
|
0.969 |
0.951 |
0.926 |
0.968 |
0.961 |
0.908 |
0.920 |
0.901 |
0.907 |
0.842 |
| fig|300852.3.peg.2040 |
0.863 |
0.845 |
0.898 |
0.963 |
0.969 |
|
0.955 |
0.914 |
0.976 |
0.958 |
0.887 |
0.877 |
0.895 |
0.889 |
0.823 |
| fig|300852.3.peg.2041 |
0.808 |
0.843 |
0.813 |
0.940 |
0.951 |
0.955 |
|
0.965 |
0.943 |
0.948 |
0.916 |
0.908 |
0.941 |
0.908 |
0.897 |
| fig|300852.3.peg.2042 |
0.814 |
0.860 |
0.734 |
0.924 |
0.926 |
0.914 |
0.965 |
|
0.913 |
0.930 |
0.899 |
0.924 |
0.929 |
0.918 |
0.924 |
| fig|300852.3.peg.2043 |
0.878 |
0.861 |
0.845 |
0.945 |
0.968 |
0.976 |
0.943 |
0.913 |
|
0.977 |
0.916 |
0.902 |
0.904 |
0.906 |
0.839 |
| fig|300852.3.peg.2044 |
0.849 |
0.854 |
0.791 |
0.939 |
0.961 |
0.958 |
0.948 |
0.930 |
0.977 |
|
0.954 |
0.936 |
0.915 |
0.928 |
0.888 |
| fig|300852.3.peg.2045 |
0.765 |
0.827 |
0.666 |
0.873 |
0.908 |
0.887 |
0.916 |
0.899 |
0.916 |
0.954 |
|
0.948 |
0.906 |
0.922 |
0.925 |
| fig|300852.3.peg.2046 |
0.824 |
0.869 |
0.663 |
0.890 |
0.920 |
0.877 |
0.908 |
0.924 |
0.902 |
0.936 |
0.948 |
|
0.908 |
0.960 |
0.938 |
| fig|300852.3.peg.2047 |
0.800 |
0.865 |
0.716 |
0.864 |
0.901 |
0.895 |
0.941 |
0.929 |
0.904 |
0.915 |
0.906 |
0.908 |
|
0.946 |
0.929 |
| fig|300852.3.peg.2048 |
0.824 |
0.882 |
0.690 |
0.875 |
0.907 |
0.889 |
0.908 |
0.918 |
0.906 |
0.928 |
0.922 |
0.960 |
0.946 |
|
0.954 |
| fig|300852.3.peg.2049 |
0.712 |
0.821 |
0.584 |
0.829 |
0.842 |
0.823 |
0.897 |
0.924 |
0.839 |
0.888 |
0.925 |
0.938 |
0.929 |
0.954 |
|
Functions in Thermus thermophilus HB8
| PEG |
Function |
Subsystems |
| fig|300852.3.peg.776 |
Vitamin B12 ABC transporter, B12-binding component BtuF |
Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.777 |
Iron(III) dicitrate transport system permease protein FecD (TC 3.A.1.14.1) |
Flavohaemoglobin |
| fig|300852.3.peg.2037 |
HoxN/HupN/NixA family cobalt transporter |
Coenzyme_B12_biosynthesis,Transport_of_Nickel_and_Cobalt |
| fig|300852.3.peg.2038 |
Cobalt-precorrin-6 synthase, anaerobic |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2039 |
Cobalt-precorrin-8x methylmutase (EC 5.4.1.2) |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2040 |
Cobalt-precorrin-6y C5-methyltransferase (EC 2.1.1.-) / Cobalt-precorrin-6y C15-methyltransferase [decarboxylating] (EC 2.1.1.-) |
Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2041 |
Cobalt-precorrin-2 C20-methyltransferase (EC 2.1.1.130) |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2042 |
Cobalt-precorrin-4 C11-methyltransferase (EC 2.1.1.133) |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2043 |
Cobalt-precorrin-3b C17-methyltransferase |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2044 |
Cobalamin biosynthesis protein CbiG |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2045 |
Sirohydrochlorin cobaltochelatase (EC 4.99.1.3) |
Coenzyme_B12_biosynthesis,Heme_and_Siroheme_Biosynthesis |
| fig|300852.3.peg.2046 |
hypothetical protein |
|
| fig|300852.3.peg.2047 |
Uroporphyrinogen-III methyltransferase (EC 2.1.1.107) |
Coenzyme_B12_biosynthesis,Dissimilatory_nitrite_reductase,Heme_and_Siroheme_Biosynthesis,Synechocystis_experimental |
| fig|300852.3.peg.2048 |
Cobalamin biosynthesis protein BluB @ 5,6-dimethylbenzimidazole synthase, flavin destructase family |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis |
| fig|300852.3.peg.2049 |
Adenosylcobinamide-phosphate synthase |
Cobalamin_synthesis,Coenzyme_B12_biosynthesis,YgfZ |
Here we are looking at two clusters of genes. Let me direct your attention to peg.2046 which
is currently uncharacterized. I would suggest from the table above that it probably plays a role
in Coenzyme B12 biosynthesis.
Here is another one that seems clear to me:
Pegs in Atomic Regulon 6 [ON=65 OFF=48]
Pearson Coefficients:
| PEG |
peg.2131 |
peg.2134 |
peg.2135 |
peg.2136 |
peg.2137 |
peg.2138 |
peg.2139 |
peg.2146 |
peg.2147 |
peg.2148 |
peg.2149 |
peg.2150 |
peg.2151 |
peg.2152 |
| fig|300852.3.peg.2131 |
|
0.778 |
0.701 |
0.752 |
0.771 |
0.823 |
0.850 |
0.446 |
0.667 |
0.517 |
0.546 |
0.628 |
0.555 |
0.538 |
| fig|300852.3.peg.2134 |
0.778 |
|
0.957 |
0.972 |
0.955 |
0.960 |
0.945 |
0.734 |
0.893 |
0.806 |
0.816 |
0.845 |
0.797 |
0.792 |
| fig|300852.3.peg.2135 |
0.701 |
0.957 |
|
0.967 |
0.939 |
0.913 |
0.882 |
0.774 |
0.909 |
0.846 |
0.875 |
0.871 |
0.835 |
0.847 |
| fig|300852.3.peg.2136 |
0.752 |
0.972 |
0.967 |
|
0.954 |
0.948 |
0.934 |
0.733 |
0.879 |
0.802 |
0.818 |
0.842 |
0.791 |
0.795 |
| fig|300852.3.peg.2137 |
0.771 |
0.955 |
0.939 |
0.954 |
|
0.958 |
0.927 |
0.648 |
0.879 |
0.805 |
0.802 |
0.821 |
0.788 |
0.768 |
| fig|300852.3.peg.2138 |
0.823 |
0.960 |
0.913 |
0.948 |
0.958 |
|
0.975 |
0.608 |
0.844 |
0.744 |
0.744 |
0.787 |
0.764 |
0.737 |
| fig|300852.3.peg.2139 |
0.850 |
0.945 |
0.882 |
0.934 |
0.927 |
0.975 |
|
0.575 |
0.807 |
0.682 |
0.685 |
0.729 |
0.693 |
0.674 |
| fig|300852.3.peg.2146 |
0.446 |
0.734 |
0.774 |
0.733 |
0.648 |
0.608 |
0.575 |
|
0.829 |
0.832 |
0.847 |
0.820 |
0.767 |
0.824 |
| fig|300852.3.peg.2147 |
0.667 |
0.893 |
0.909 |
0.879 |
0.879 |
0.844 |
0.807 |
0.829 |
|
0.952 |
0.960 |
0.955 |
0.941 |
0.945 |
| fig|300852.3.peg.2148 |
0.517 |
0.806 |
0.846 |
0.802 |
0.805 |
0.744 |
0.682 |
0.832 |
0.952 |
|
0.953 |
0.923 |
0.945 |
0.941 |
| fig|300852.3.peg.2149 |
0.546 |
0.816 |
0.875 |
0.818 |
0.802 |
0.744 |
0.685 |
0.847 |
0.960 |
0.953 |
|
0.971 |
0.953 |
0.974 |
| fig|300852.3.peg.2150 |
0.628 |
0.845 |
0.871 |
0.842 |
0.821 |
0.787 |
0.729 |
0.820 |
0.955 |
0.923 |
0.971 |
|
0.959 |
0.970 |
| fig|300852.3.peg.2151 |
0.555 |
0.797 |
0.835 |
0.791 |
0.788 |
0.764 |
0.693 |
0.767 |
0.941 |
0.945 |
0.953 |
0.959 |
|
0.970 |
| fig|300852.3.peg.2152 |
0.538 |
0.792 |
0.847 |
0.795 |
0.768 |
0.737 |
0.674 |
0.824 |
0.945 |
0.941 |
0.974 |
0.970 |
0.970 |
|
Functions in Thermus thermophilus HB8
| PEG |
Function |
Subsystems |
| fig|300852.3.peg.2131 |
CRISPR-associated protein Cas02710 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2134 |
CRISPR-associated protein, Csm1 family |
CRISPR-associated_cluster |
| fig|300852.3.peg.2135 |
CRISPR-associated protein, Csm2 family |
CRISPR-associated_cluster |
| fig|300852.3.peg.2136 |
CRISPR-associated RAMP Csm3 |
CRISPR-associated_cluster |
| fig|300852.3.peg.2137 |
CRISPR-associated RAMP protein, Csm4 family |
CRISPR-associated_cluster |
| fig|300852.3.peg.2138 |
CRISPR-associated protein, Csm5 family |
CRISPR-associated_cluster |
| fig|300852.3.peg.2139 |
CRISPR-associated protein Cas02710 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2146 |
hypothetical protein |
|
| fig|300852.3.peg.2147 |
CRISPR-associated RAMP Cmr2 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2148 |
CRISPR-associated RAMP Cmr3 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2149 |
CRISPR-associated RAMP Cmr1 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2150 |
CRISPR-associated RAMP Cmr4 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2151 |
CRISPR-associated RAMP Cmr5 |
CRISP_Cmr_Cluster |
| fig|300852.3.peg.2152 |
CRISPR-associated RAMP Cmr6 |
CRISP_Cmr_Cluster |
The hypothetical peg.2146 is not in an operon with the other CRISPR-associated proteins; it
is divergent. Without the expression data, it would be hard to be certain. With it, it
seems pretty clear (I think!). Note that expression is ON in 65 experiments and OFF in 48.
Well, that illustrates my point that the atomic regulons are obviously a powerful source of
conjectures relating to the functions of gene products. I have selected most of my examples
from Escherichia coli because I think that it will make it easier for the reader to evaluate their
significane; however, it should be emphasized that it is completely straightforward to generate
reasonable conjectures for a number of organisms already.
Pegs in Atomic Regulon 2 [ON=139 OFF=711]
Pearson Coefficients:
| PEG |
peg.848 |
peg.849 |
peg.859 |
peg.862 |
peg.880 |
peg.885 |
peg.886 |
peg.887 |
peg.888 |
peg.889 |
peg.890 |
peg.891 |
peg.892 |
peg.893 |
peg.894 |
peg.895 |
peg.896 |
peg.897 |
peg.898 |
peg.899 |
peg.900 |
peg.901 |
peg.902 |
peg.903 |
peg.904 |
peg.905 |
peg.906 |
peg.907 |
peg.909 |
peg.910 |
peg.911 |
peg.2753 |
| fig|158878.1.peg.848 |
|
0.920 |
0.745 |
0.793 |
0.751 |
0.760 |
0.723 |
0.758 |
0.737 |
0.719 |
0.794 |
0.763 |
0.783 |
0.766 |
0.780 |
0.790 |
0.786 |
0.766 |
0.723 |
0.593 |
0.742 |
0.761 |
0.729 |
0.655 |
0.639 |
0.687 |
0.723 |
0.712 |
0.638 |
0.713 |
0.711 |
0.738 |
| fig|158878.1.peg.849 |
0.920 |
|
0.713 |
0.792 |
0.787 |
0.741 |
0.707 |
0.738 |
0.707 |
0.683 |
0.798 |
0.755 |
0.776 |
0.748 |
0.761 |
0.793 |
0.780 |
0.753 |
0.708 |
0.538 |
0.718 |
0.743 |
0.709 |
0.611 |
0.637 |
0.667 |
0.715 |
0.684 |
0.628 |
0.713 |
0.711 |
0.739 |
| fig|158878.1.peg.859 |
0.745 |
0.713 |
|
0.736 |
0.593 |
0.714 |
0.679 |
0.696 |
0.667 |
0.702 |
0.684 |
0.690 |
0.682 |
0.667 |
0.681 |
0.698 |
0.697 |
0.693 |
0.638 |
0.534 |
0.652 |
0.682 |
0.657 |
0.591 |
0.627 |
0.669 |
0.689 |
0.690 |
0.603 |
0.713 |
0.710 |
0.820 |
| fig|158878.1.peg.862 |
0.793 |
0.792 |
0.736 |
|
0.712 |
0.711 |
0.684 |
0.696 |
0.654 |
0.675 |
0.715 |
0.728 |
0.695 |
0.704 |
0.681 |
0.721 |
0.713 |
0.712 |
0.626 |
0.527 |
0.644 |
0.693 |
0.657 |
0.578 |
0.590 |
0.623 |
0.624 |
0.656 |
0.600 |
0.653 |
0.666 |
0.734 |
| fig|158878.1.peg.880 |
0.751 |
0.787 |
0.593 |
0.712 |
|
0.677 |
0.634 |
0.662 |
0.623 |
0.638 |
0.640 |
0.688 |
0.655 |
0.673 |
0.663 |
0.696 |
0.668 |
0.694 |
0.631 |
0.542 |
0.639 |
0.712 |
0.651 |
0.580 |
0.586 |
0.636 |
0.616 |
0.630 |
0.611 |
0.663 |
0.669 |
0.625 |
| fig|158878.1.peg.885 |
0.760 |
0.741 |
0.714 |
0.711 |
0.677 |
|
0.909 |
0.854 |
0.813 |
0.820 |
0.805 |
0.862 |
0.837 |
0.835 |
0.851 |
0.829 |
0.846 |
0.865 |
0.807 |
0.689 |
0.812 |
0.852 |
0.818 |
0.744 |
0.751 |
0.708 |
0.734 |
0.785 |
0.767 |
0.784 |
0.783 |
0.691 |
| fig|158878.1.peg.886 |
0.723 |
0.707 |
0.679 |
0.684 |
0.634 |
0.909 |
|
0.883 |
0.848 |
0.780 |
0.802 |
0.877 |
0.821 |
0.836 |
0.864 |
0.786 |
0.832 |
0.874 |
0.833 |
0.774 |
0.815 |
0.868 |
0.826 |
0.812 |
0.825 |
0.763 |
0.721 |
0.764 |
0.831 |
0.776 |
0.790 |
0.655 |
| fig|158878.1.peg.887 |
0.758 |
0.738 |
0.696 |
0.696 |
0.662 |
0.854 |
0.883 |
|
0.918 |
0.851 |
0.826 |
0.897 |
0.862 |
0.894 |
0.915 |
0.848 |
0.911 |
0.895 |
0.926 |
0.837 |
0.906 |
0.938 |
0.929 |
0.893 |
0.840 |
0.844 |
0.811 |
0.828 |
0.889 |
0.863 |
0.864 |
0.727 |
| fig|158878.1.peg.888 |
0.737 |
0.707 |
0.667 |
0.654 |
0.623 |
0.813 |
0.848 |
0.918 |
|
0.824 |
0.842 |
0.867 |
0.857 |
0.866 |
0.918 |
0.830 |
0.892 |
0.874 |
0.928 |
0.865 |
0.909 |
0.909 |
0.923 |
0.913 |
0.836 |
0.834 |
0.856 |
0.834 |
0.863 |
0.832 |
0.830 |
0.707 |
| fig|158878.1.peg.889 |
0.719 |
0.683 |
0.702 |
0.675 |
0.638 |
0.820 |
0.780 |
0.851 |
0.824 |
|
0.822 |
0.847 |
0.808 |
0.845 |
0.824 |
0.804 |
0.859 |
0.846 |
0.819 |
0.707 |
0.829 |
0.864 |
0.845 |
0.751 |
0.702 |
0.737 |
0.743 |
0.844 |
0.756 |
0.774 |
0.787 |
0.707 |
| fig|158878.1.peg.890 |
0.794 |
0.798 |
0.684 |
0.715 |
0.640 |
0.805 |
0.802 |
0.826 |
0.842 |
0.822 |
|
0.887 |
0.883 |
0.887 |
0.859 |
0.847 |
0.887 |
0.891 |
0.843 |
0.670 |
0.840 |
0.831 |
0.833 |
0.750 |
0.768 |
0.751 |
0.827 |
0.861 |
0.763 |
0.781 |
0.783 |
0.739 |
| fig|158878.1.peg.891 |
0.763 |
0.755 |
0.690 |
0.728 |
0.688 |
0.862 |
0.877 |
0.897 |
0.867 |
0.847 |
0.887 |
|
0.847 |
0.901 |
0.884 |
0.837 |
0.902 |
0.942 |
0.888 |
0.806 |
0.869 |
0.921 |
0.875 |
0.842 |
0.854 |
0.829 |
0.777 |
0.856 |
0.876 |
0.815 |
0.847 |
0.709 |
| fig|158878.1.peg.892 |
0.783 |
0.776 |
0.682 |
0.695 |
0.655 |
0.837 |
0.821 |
0.862 |
0.857 |
0.808 |
0.883 |
0.847 |
|
0.892 |
0.909 |
0.880 |
0.911 |
0.874 |
0.873 |
0.679 |
0.872 |
0.852 |
0.875 |
0.768 |
0.726 |
0.725 |
0.848 |
0.818 |
0.738 |
0.819 |
0.796 |
0.729 |
| fig|158878.1.peg.893 |
0.766 |
0.748 |
0.667 |
0.704 |
0.673 |
0.835 |
0.836 |
0.894 |
0.866 |
0.845 |
0.887 |
0.901 |
0.892 |
|
0.906 |
0.869 |
0.920 |
0.919 |
0.901 |
0.761 |
0.893 |
0.902 |
0.899 |
0.816 |
0.787 |
0.780 |
0.823 |
0.863 |
0.816 |
0.835 |
0.830 |
0.720 |
| fig|158878.1.peg.894 |
0.780 |
0.761 |
0.681 |
0.681 |
0.663 |
0.851 |
0.864 |
0.915 |
0.918 |
0.824 |
0.859 |
0.884 |
0.909 |
0.906 |
|
0.861 |
0.922 |
0.908 |
0.932 |
0.808 |
0.919 |
0.912 |
0.916 |
0.870 |
0.807 |
0.808 |
0.867 |
0.831 |
0.825 |
0.848 |
0.841 |
0.731 |
| fig|158878.1.peg.895 |
0.790 |
0.793 |
0.698 |
0.721 |
0.696 |
0.829 |
0.786 |
0.848 |
0.830 |
0.804 |
0.847 |
0.837 |
0.880 |
0.869 |
0.861 |
|
0.900 |
0.856 |
0.841 |
0.661 |
0.840 |
0.840 |
0.863 |
0.739 |
0.705 |
0.721 |
0.819 |
0.821 |
0.740 |
0.814 |
0.811 |
0.722 |
| fig|158878.1.peg.896 |
0.786 |
0.780 |
0.697 |
0.713 |
0.668 |
0.846 |
0.832 |
0.911 |
0.892 |
0.859 |
0.887 |
0.902 |
0.911 |
0.920 |
0.922 |
0.900 |
|
0.910 |
0.915 |
0.749 |
0.910 |
0.915 |
0.922 |
0.824 |
0.779 |
0.802 |
0.858 |
0.866 |
0.815 |
0.860 |
0.848 |
0.755 |
| fig|158878.1.peg.897 |
0.766 |
0.753 |
0.693 |
0.712 |
0.694 |
0.865 |
0.874 |
0.895 |
0.874 |
0.846 |
0.891 |
0.942 |
0.874 |
0.919 |
0.908 |
0.856 |
0.910 |
|
0.897 |
0.804 |
0.880 |
0.918 |
0.885 |
0.838 |
0.833 |
0.806 |
0.811 |
0.869 |
0.848 |
0.820 |
0.846 |
0.708 |
| fig|158878.1.peg.898 |
0.723 |
0.708 |
0.638 |
0.626 |
0.631 |
0.807 |
0.833 |
0.926 |
0.928 |
0.819 |
0.843 |
0.888 |
0.873 |
0.901 |
0.932 |
0.841 |
0.915 |
0.897 |
|
0.858 |
0.925 |
0.936 |
0.941 |
0.918 |
0.832 |
0.852 |
0.853 |
0.845 |
0.879 |
0.862 |
0.860 |
0.707 |
| fig|158878.1.peg.899 |
0.593 |
0.538 |
0.534 |
0.527 |
0.542 |
0.689 |
0.774 |
0.837 |
0.865 |
0.707 |
0.670 |
0.806 |
0.679 |
0.761 |
0.808 |
0.661 |
0.749 |
0.804 |
0.858 |
|
0.814 |
0.877 |
0.829 |
0.941 |
0.838 |
0.836 |
0.701 |
0.735 |
0.887 |
0.731 |
0.770 |
0.564 |
| fig|158878.1.peg.900 |
0.742 |
0.718 |
0.652 |
0.644 |
0.639 |
0.812 |
0.815 |
0.906 |
0.909 |
0.829 |
0.840 |
0.869 |
0.872 |
0.893 |
0.919 |
0.840 |
0.910 |
0.880 |
0.925 |
0.814 |
|
0.918 |
0.919 |
0.872 |
0.792 |
0.810 |
0.856 |
0.840 |
0.828 |
0.843 |
0.833 |
0.722 |
| fig|158878.1.peg.901 |
0.761 |
0.743 |
0.682 |
0.693 |
0.712 |
0.852 |
0.868 |
0.938 |
0.909 |
0.864 |
0.831 |
0.921 |
0.852 |
0.902 |
0.912 |
0.840 |
0.915 |
0.918 |
0.936 |
0.877 |
0.918 |
|
0.935 |
0.906 |
0.848 |
0.875 |
0.811 |
0.862 |
0.905 |
0.864 |
0.875 |
0.728 |
| fig|158878.1.peg.902 |
0.729 |
0.709 |
0.657 |
0.657 |
0.651 |
0.818 |
0.826 |
0.929 |
0.923 |
0.845 |
0.833 |
0.875 |
0.875 |
0.899 |
0.916 |
0.863 |
0.922 |
0.885 |
0.941 |
0.829 |
0.919 |
0.935 |
|
0.891 |
0.808 |
0.841 |
0.854 |
0.857 |
0.864 |
0.874 |
0.861 |
0.729 |
| fig|158878.1.peg.903 |
0.655 |
0.611 |
0.591 |
0.578 |
0.580 |
0.744 |
0.812 |
0.893 |
0.913 |
0.751 |
0.750 |
0.842 |
0.768 |
0.816 |
0.870 |
0.739 |
0.824 |
0.838 |
0.918 |
0.941 |
0.872 |
0.906 |
0.891 |
|
0.868 |
0.866 |
0.804 |
0.789 |
0.908 |
0.817 |
0.827 |
0.645 |
| fig|158878.1.peg.904 |
0.639 |
0.637 |
0.627 |
0.590 |
0.586 |
0.751 |
0.825 |
0.840 |
0.836 |
0.702 |
0.768 |
0.854 |
0.726 |
0.787 |
0.807 |
0.705 |
0.779 |
0.833 |
0.832 |
0.838 |
0.792 |
0.848 |
0.808 |
0.868 |
|
0.881 |
0.788 |
0.793 |
0.891 |
0.840 |
0.870 |
0.707 |
| fig|158878.1.peg.905 |
0.687 |
0.667 |
0.669 |
0.623 |
0.636 |
0.708 |
0.763 |
0.844 |
0.834 |
0.737 |
0.751 |
0.829 |
0.725 |
0.780 |
0.808 |
0.721 |
0.802 |
0.806 |
0.852 |
0.836 |
0.810 |
0.875 |
0.841 |
0.866 |
0.881 |
|
0.805 |
0.785 |
0.869 |
0.866 |
0.882 |
0.750 |
| fig|158878.1.peg.906 |
0.723 |
0.715 |
0.689 |
0.624 |
0.616 |
0.734 |
0.721 |
0.811 |
0.856 |
0.743 |
0.827 |
0.777 |
0.848 |
0.823 |
0.867 |
0.819 |
0.858 |
0.811 |
0.853 |
0.701 |
0.856 |
0.811 |
0.854 |
0.804 |
0.788 |
0.805 |
|
0.839 |
0.738 |
0.872 |
0.842 |
0.776 |
| fig|158878.1.peg.907 |
0.712 |
0.684 |
0.690 |
0.656 |
0.630 |
0.785 |
0.764 |
0.828 |
0.834 |
0.844 |
0.861 |
0.856 |
0.818 |
0.863 |
0.831 |
0.821 |
0.866 |
0.869 |
0.845 |
0.735 |
0.840 |
0.862 |
0.857 |
0.789 |
0.793 |
0.785 |
0.839 |
|
0.810 |
0.832 |
0.835 |
0.745 |
| fig|158878.1.peg.909 |
0.638 |
0.628 |
0.603 |
0.600 |
0.611 |
0.767 |
0.831 |
0.889 |
0.863 |
0.756 |
0.763 |
0.876 |
0.738 |
0.816 |
0.825 |
0.740 |
0.815 |
0.848 |
0.879 |
0.887 |
0.828 |
0.905 |
0.864 |
0.908 |
0.891 |
0.869 |
0.738 |
0.810 |
|
0.817 |
0.839 |
0.653 |
| fig|158878.1.peg.910 |
0.713 |
0.713 |
0.713 |
0.653 |
0.663 |
0.784 |
0.776 |
0.863 |
0.832 |
0.774 |
0.781 |
0.815 |
0.819 |
0.835 |
0.848 |
0.814 |
0.860 |
0.820 |
0.862 |
0.731 |
0.843 |
0.864 |
0.874 |
0.817 |
0.840 |
0.866 |
0.872 |
0.832 |
0.817 |
|
0.937 |
0.823 |
| fig|158878.1.peg.911 |
0.711 |
0.711 |
0.710 |
0.666 |
0.669 |
0.783 |
0.790 |
0.864 |
0.830 |
0.787 |
0.783 |
0.847 |
0.796 |
0.830 |
0.841 |
0.811 |
0.848 |
0.846 |
0.860 |
0.770 |
0.833 |
0.875 |
0.861 |
0.827 |
0.870 |
0.882 |
0.842 |
0.835 |
0.839 |
0.937 |
|
0.816 |
| fig|158878.1.peg.2753 |
0.738 |
0.739 |
0.820 |
0.734 |
0.625 |
0.691 |
0.655 |
0.727 |
0.707 |
0.707 |
0.739 |
0.709 |
0.729 |
0.720 |
0.731 |
0.722 |
0.755 |
0.708 |
0.707 |
0.564 |
0.722 |
0.728 |
0.729 |
0.645 |
0.707 |
0.750 |
0.776 |
0.745 |
0.653 |
0.823 |
0.816 |
|
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.848 |
Excisionase [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.849 |
Hypothetical protein, SAV0849 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.859 |
Hypothetical protein, PV83 orf12 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.862 |
Hypothetical protein, phi-ETA orf16 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.880 |
Hypothetical protein, SAV0880 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.885 |
Phage terminase, small subunit |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.886 |
Phage terminase, large subunit |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.887 |
Phage portal |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.888 |
Phage minor head protein |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.889 |
unknown function |
|
| fig|158878.1.peg.890 |
Phage capsid and scaffold |
Phage_capsid_proteins,Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.891 |
Phage head protein [SA bacteriophages 11, Mu50B] / Phage major capsid protein #Fam0008 |
Phage_capsid_proteins,Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.892 |
Phage major capsid protein #Fam0008 |
Phage_capsid_proteins,Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.893 |
Phage transcriptional terminator |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.894 |
phi 11 orf36 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.895 |
hypothetical protein |
|
| fig|158878.1.peg.896 |
phi 11 orf37 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.897 |
phi 11 orf38 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.898 |
Phage tail protein |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.899 |
phi 11 orf40 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.900 |
phi 11 orf41 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.901 |
Tape measure protein [SA bacteriophages 11, Mu50B] |
Phage_tail_proteins,Phage_tail_proteins_2,Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.902 |
phi 11 orf43 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.903 |
Phage minor structural protein |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.904 |
Putative major teichoic acid biosynthesis protein C |
Staphylococcal_phi-Mu50B-like_prophages,Teichoic_and_lipoteichoic_acids_biosynthesis |
| fig|158878.1.peg.905 |
hypothetical protein within a prophage |
|
| fig|158878.1.peg.906 |
Hypothetical protein, phi-ETA orf42 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.907 |
Hypothetical protein, phi-ETA orf58 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.909 |
Bifunctional autolysin Atl / N-acetylmuramoyl-L-alanine amidase (EC 3.5.1.28) / Endo-beta-N-acetylglucosaminidase (EC 3.2.1.96) |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.910 |
Tail fiber protein [SA bacteriophages 11, Mu50B] [SS] |
Phage_tail_fiber_proteins,Phage_tail_proteins,Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.911 |
Hypothetical protein, phi-ETA orf63 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| fig|158878.1.peg.2753 |
Phage protein |
Staphylococcal_phi-Mu50B-like_prophages |
The genes encoding peg.890 or peg.895 are clearly implicated in phage-related machinery.
The context supplied by the atomic regulon makes this apparent.
Pegs in Atomic Regulon 3 [ON=390 OFF=451]
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.2388 |
Nitrate/nitrite transporter |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2391 |
Two-component response regulator |
|
| fig|158878.1.peg.2392 |
Two component sensor histidine kinase |
|
| fig|158878.1.peg.2393 |
hypothetical protein |
|
| fig|158878.1.peg.2394 |
Respiratory nitrate reductase gamma chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2395 |
Respiratory nitrate reductase delta chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2396 |
Respiratory nitrate reductase beta chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2397 |
Respiratory nitrate reductase alpha chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2398 |
Uroporphyrinogen-III methyltransferase (EC 2.1.1.107) |
Coenzyme_B12_biosynthesis,Dissimilatory_nitrite_reductase,Heme_and_Siroheme_Biosynthesis,Synechocystis_experimental |
| fig|158878.1.peg.2399 |
Nitrite reductase [NAD(P)H] small subunit (EC 1.7.1.4) |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2400 |
Nitrite reductase [NAD(P)H] large subunit (EC 1.7.1.4) |
Nitrate_and_nitrite_ammonification |
| fig|158878.1.peg.2401 |
Sirohydrochlorin ferrochelatase (EC 4.99.1.4) |
Heme_and_Siroheme_Biosynthesis |
The connection between peg.2388, peg.2391, peg.2392, peg.2401 and
the rest of the cluster appears questionable. In such cases, it is useful to examine
the strengths of the Pearson Correlation coefficients (PCs) against all of the other genes
in the genome. Here are the relevant summaries:
Connections for fig|158878.1.peg.2388
| PC |
PEG |
Function |
Subsystems |
| 0.815 |
fig|158878.1.peg.2400 |
Nitrite reductase [NAD(P)H] large subunit (EC 1.7.1.4) |
Nitrate_and_nitrite_ammonification |
| 0.799 |
fig|158878.1.peg.2398 |
Uroporphyrinogen-III methyltransferase (EC 2.1.1.107) |
Coenzyme_B12_biosynthesis,Dissimilatory_nitrite_reductase,Heme_and_Siroheme_Biosynthesis,Synechocystis_experimental |
| 0.790 |
fig|158878.1.peg.2399 |
Nitrite reductase [NAD(P)H] small subunit (EC 1.7.1.4) |
Nitrate_and_nitrite_ammonification |
| 0.759 |
fig|158878.1.peg.2401 |
Sirohydrochlorin ferrochelatase (EC 4.99.1.4) |
Heme_and_Siroheme_Biosynthesis |
| 0.753 |
fig|158878.1.peg.2397 |
Respiratory nitrate reductase alpha chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| 0.745 |
fig|158878.1.peg.2396 |
Respiratory nitrate reductase beta chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| 0.737 |
fig|158878.1.peg.2395 |
Respiratory nitrate reductase delta chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
Connections for fig|158878.1.peg.2401
| PC |
PEG |
Function |
Subsystems |
| 0.858 |
fig|158878.1.peg.2398 |
Uroporphyrinogen-III methyltransferase (EC 2.1.1.107) |
Coenzyme_B12_biosynthesis,Dissimilatory_nitrite_reductase,Heme_and_Siroheme_Biosynthesis,Synechocystis_experimental |
| 0.847 |
fig|158878.1.peg.2396 |
Respiratory nitrate reductase beta chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| 0.842 |
fig|158878.1.peg.2400 |
Nitrite reductase [NAD(P)H] large subunit (EC 1.7.1.4) |
Nitrate_and_nitrite_ammonification |
| 0.841 |
fig|158878.1.peg.2397 |
Respiratory nitrate reductase alpha chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| 0.840 |
fig|158878.1.peg.2399 |
Nitrite reductase [NAD(P)H] small subunit (EC 1.7.1.4) |
Nitrate_and_nitrite_ammonification |
| 0.785 |
fig|158878.1.peg.305 |
formate/nitrite transporter family protein |
|
| 0.769 |
fig|158878.1.peg.2395 |
Respiratory nitrate reductase delta chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
| 0.759 |
fig|158878.1.peg.2388 |
Nitrate/nitrite transporter |
Nitrate_and_nitrite_ammonification |
| 0.721 |
fig|158878.1.peg.2394 |
Respiratory nitrate reductase gamma chain (EC 1.7.99.4) |
Nitrate_and_nitrite_ammonification |
In each case, I have shown just the best hits (i.e., I deleted only weaker hits than those
shown). This, in my mind, makes an interesting case that
-
the two-component regulatory system implemented by peg.2391 and peg.2392
relate to "Nitrate and nitrite ammonification" (I am not even sure what that precisely means, but I think that
it is straightforward to study the subsystem and literature to gain a detailed grasp), and
-
peg.2393 relates to "Nitrate and nitrite ammonification" or to the regulation of that process.
Pegs in Atomic Regulon 12 [ON=97 OFF=751]
This next example appears to quite problematic. It illustrates how a combination of clues
can be accumulated, gradually leading to a situation in which meaningful
conjectures might be formulated.
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.399 |
hypothetical protein |
|
| fig|158878.1.peg.400 |
lipoprotein, NLP/P60 family |
|
| fig|158878.1.peg.401 |
hypothetical protein |
|
| fig|158878.1.peg.402 |
hypothetical protein |
|
| fig|158878.1.peg.403 |
hypothetical protein |
|
| fig|158878.1.peg.404 |
hypothetical protein |
|
| fig|158878.1.peg.405 |
hypothetical protein |
|
| fig|158878.1.peg.406 |
hypothetical protein |
|
| fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
First, it is worth noting that very similar clusters of genes exist in Listeria monocytogenes EGD-e,
Enterococcus faecalis V583, Enterococcus faecalis plasmid pCF10, Clostridium difficile,
Streptococcus pneumoniae, and Streptococcus agalactiae. It is the result of a plasmid
conjugation event (I think).
A few related facts support this hypothesis:
- peg.400 has a hit against a domain described as "invasion associated secreted endopeptidase".
- peg.401 hits a domain described as "Intracellular trafficking and secretion".
- peg.402 hits a domain described by TIGR02746 as
type-IV secretion system protein TraC The protein family
described here is common among the F, P and I-like type IV secretion
systems. Gene symbols include TraC (F-type), TrbE/VirB4 (P-type) and
TraU (I-type). The protein conyains the Walker A and B motifs and so
is a putative nucleotide triphosphatase.
- peg.403 hits a domain described as
Antirestriction protein (ArdA)
This family consists of several bacterial antirestriction (ArdA)
proteins. ArdA functions in bacterial conjugation to allow an
unmodified plasmid to evade restriction in the recipient bacterium and
yet acquire cognate modification.
- a FIG annotator assigned to peg.404 the function "FIG086557: Conjugation related protein"
Taken together these paint an obvious picture, I think.
Now, if we look at the precise PC connections for some of the genes, we see
Connections for fig|158878.1.peg.400
| PC |
PEG |
Function |
Subsystems |
| 0.932 |
fig|158878.1.peg.402 |
hypothetical protein |
|
| 0.925 |
fig|158878.1.peg.408 |
hypothetical protein |
|
| 0.925 |
fig|158878.1.peg.409 |
hypothetical protein |
|
| 0.924 |
fig|158878.1.peg.411 |
hypothetical protein |
|
| 0.924 |
fig|158878.1.peg.405 |
hypothetical protein |
|
| 0.913 |
fig|158878.1.peg.401 |
hypothetical protein |
|
| 0.905 |
fig|158878.1.peg.415 |
Transposase |
|
| 0.902 |
fig|158878.1.peg.412 |
hypothetical protein |
|
| 0.901 |
fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
| 0.898 |
fig|158878.1.peg.792 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.889 |
fig|158878.1.peg.403 |
hypothetical protein |
|
| 0.885 |
fig|158878.1.peg.398 |
Tetracycline resistance protein TetM |
Tetracycline_resistance,_ribosome_protection_type,Translation_elongation_factor_G_family |
| 0.884 |
fig|158878.1.peg.410 |
hypothetical protein |
|
| 0.869 |
fig|158878.1.peg.865 |
HNH homing endonuclease |
|
| 0.859 |
fig|158878.1.peg.399 |
hypothetical protein |
|
| 0.858 |
fig|158878.1.peg.394 |
hhypothetical protein |
|
| 0.853 |
fig|158878.1.peg.791 |
Hypothetical SAV0791 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.851 |
fig|158878.1.peg.868 |
Phage replication initiation |
|
| 0.843 |
fig|158878.1.peg.397 |
hypothetical protein |
|
| 0.842 |
fig|158878.1.peg.406 |
hypothetical protein |
|
| 0.841 |
fig|158878.1.peg.1989 |
hypothetical protein within prophage phiN315 |
|
| 0.829 |
fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| 0.829 |
fig|158878.1.peg.392 |
Integrase |
|
| 0.823 |
fig|158878.1.peg.395 |
hypothetical protein |
|
| 0.823 |
fig|158878.1.peg.916 |
hypothetical protein |
|
| 0.821 |
fig|158878.1.peg.404 |
hypothetical protein |
|
| 0.820 |
fig|158878.1.peg.858 |
unknown function |
|
| 0.818 |
fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| 0.796 |
fig|158878.1.peg.854 |
unknown function |
|
| 0.787 |
fig|158878.1.peg.2021 |
Putative DNA helicase, superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.786 |
fig|158878.1.peg.866 |
Hypothetical protein, PV83 orf19 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.774 |
fig|158878.1.peg.869 |
DNA replication protein DnaC |
DNA-replication |
| 0.765 |
fig|158878.1.peg.912 |
Phage holin |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| 0.757 |
fig|158878.1.peg.2020 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.752 |
fig|158878.1.peg.1999 |
hypothetical protein |
|
| 0.751 |
fig|158878.1.peg.790 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.750 |
fig|158878.1.peg.47 |
Disulfide bond regulator |
Disulphide_related_cluster |
| 0.750 |
fig|158878.1.peg.884 |
Integrase regulator RinA |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.746 |
fig|158878.1.peg.872 |
Hypothetical protein, PV83 orf22 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.739 |
fig|158878.1.peg.2023 |
Hypothetical SAV0789 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.737 |
fig|158878.1.peg.72 |
Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1) |
Potassium_homeostasis |
| 0.726 |
fig|158878.1.peg.32 |
hypothetical protein |
|
| 0.720 |
fig|158878.1.peg.76 |
hypothetical protein |
|
| 0.718 |
fig|158878.1.peg.34 |
Bleomycin resistance protein |
|
| 0.715 |
fig|158878.1.peg.71 |
Osmosensitive K+ channel histidine kinase KdpD (EC 2.7.3.-) |
Potassium_homeostasis |
| 0.708 |
fig|158878.1.peg.802 |
Pathogenicity island SaPIn1 |
|
| 0.707 |
fig|158878.1.peg.49 |
FIG003846: hypothetical protein |
Disulphide_related_cluster |
| 0.703 |
fig|158878.1.peg.69 |
Conserved hypothetical protein |
|
| 0.701 |
fig|158878.1.peg.2022 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
Connections for fig|158878.1.peg.401
| PC |
PEG |
Function |
Subsystems |
| 0.913 |
fig|158878.1.peg.409 |
hypothetical protein |
|
| 0.913 |
fig|158878.1.peg.400 |
lipoprotein, NLP/P60 family |
|
| 0.911 |
fig|158878.1.peg.402 |
hypothetical protein |
|
| 0.904 |
fig|158878.1.peg.411 |
hypothetical protein |
|
| 0.900 |
fig|158878.1.peg.412 |
hypothetical protein |
|
| 0.899 |
fig|158878.1.peg.405 |
hypothetical protein |
|
| 0.892 |
fig|158878.1.peg.408 |
hypothetical protein |
|
| 0.890 |
fig|158878.1.peg.415 |
Transposase |
|
| 0.870 |
fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
| 0.860 |
fig|158878.1.peg.398 |
Tetracycline resistance protein TetM |
Tetracycline_resistance,_ribosome_protection_type,Translation_elongation_factor_G_family |
| 0.847 |
fig|158878.1.peg.394 |
hhypothetical protein |
|
| 0.844 |
fig|158878.1.peg.792 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.841 |
fig|158878.1.peg.399 |
hypothetical protein |
|
| 0.839 |
fig|158878.1.peg.410 |
hypothetical protein |
|
| 0.832 |
fig|158878.1.peg.403 |
hypothetical protein |
|
| 0.821 |
fig|158878.1.peg.392 |
Integrase |
|
| 0.820 |
fig|158878.1.peg.865 |
HNH homing endonuclease |
|
| 0.820 |
fig|158878.1.peg.791 |
Hypothetical SAV0791 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.816 |
fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| 0.816 |
fig|158878.1.peg.406 |
hypothetical protein |
|
| 0.815 |
fig|158878.1.peg.397 |
hypothetical protein |
|
| 0.805 |
fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| 0.803 |
fig|158878.1.peg.868 |
Phage replication initiation |
|
| 0.787 |
fig|158878.1.peg.404 |
hypothetical protein |
|
| 0.783 |
fig|158878.1.peg.1989 |
hypothetical protein within prophage phiN315 |
|
| 0.782 |
fig|158878.1.peg.869 |
DNA replication protein DnaC |
DNA-replication |
| 0.778 |
fig|158878.1.peg.395 |
hypothetical protein |
|
| 0.768 |
fig|158878.1.peg.866 |
Hypothetical protein, PV83 orf19 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.761 |
fig|158878.1.peg.916 |
hypothetical protein |
|
| 0.747 |
fig|158878.1.peg.2021 |
Putative DNA helicase, superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.742 |
fig|158878.1.peg.858 |
unknown function |
|
| 0.728 |
fig|158878.1.peg.2020 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.723 |
fig|158878.1.peg.912 |
Phage holin |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| 0.721 |
fig|158878.1.peg.790 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.720 |
fig|158878.1.peg.884 |
Integrase regulator RinA |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.711 |
fig|158878.1.peg.47 |
Disulfide bond regulator |
Disulphide_related_cluster |
| 0.709 |
fig|158878.1.peg.872 |
Hypothetical protein, PV83 orf22 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.706 |
fig|158878.1.peg.72 |
Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1) |
Potassium_homeostasis |
Connections for fig|158878.1.peg.402
| PC |
PEG |
Function |
Subsystems |
| 0.937 |
fig|158878.1.peg.405 |
hypothetical protein |
|
| 0.932 |
fig|158878.1.peg.400 |
lipoprotein, NLP/P60 family |
|
| 0.925 |
fig|158878.1.peg.411 |
hypothetical protein |
|
| 0.924 |
fig|158878.1.peg.408 |
hypothetical protein |
|
| 0.914 |
fig|158878.1.peg.409 |
hypothetical protein |
|
| 0.911 |
fig|158878.1.peg.401 |
hypothetical protein |
|
| 0.905 |
fig|158878.1.peg.415 |
Transposase |
|
| 0.903 |
fig|158878.1.peg.412 |
hypothetical protein |
|
| 0.903 |
fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
| 0.895 |
fig|158878.1.peg.398 |
Tetracycline resistance protein TetM |
Tetracycline_resistance,_ribosome_protection_type,Translation_elongation_factor_G_family |
| 0.878 |
fig|158878.1.peg.410 |
hypothetical protein |
|
| 0.878 |
fig|158878.1.peg.403 |
hypothetical protein |
|
| 0.878 |
fig|158878.1.peg.399 |
hypothetical protein |
|
| 0.878 |
fig|158878.1.peg.792 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.869 |
fig|158878.1.peg.394 |
hhypothetical protein |
|
| 0.851 |
fig|158878.1.peg.868 |
Phage replication initiation |
|
| 0.849 |
fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| 0.849 |
fig|158878.1.peg.865 |
HNH homing endonuclease |
|
| 0.849 |
fig|158878.1.peg.791 |
Hypothetical SAV0791 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.842 |
fig|158878.1.peg.406 |
hypothetical protein |
|
| 0.840 |
fig|158878.1.peg.397 |
hypothetical protein |
|
| 0.832 |
fig|158878.1.peg.392 |
Integrase |
|
| 0.829 |
fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| 0.827 |
fig|158878.1.peg.404 |
hypothetical protein |
|
| 0.824 |
fig|158878.1.peg.395 |
hypothetical protein |
|
| 0.813 |
fig|158878.1.peg.1989 |
hypothetical protein within prophage phiN315 |
|
| 0.811 |
fig|158878.1.peg.858 |
unknown function |
|
| 0.788 |
fig|158878.1.peg.2021 |
Putative DNA helicase, superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.787 |
fig|158878.1.peg.916 |
hypothetical protein |
|
| 0.770 |
fig|158878.1.peg.869 |
DNA replication protein DnaC |
DNA-replication |
| 0.769 |
fig|158878.1.peg.854 |
unknown function |
|
| 0.765 |
fig|158878.1.peg.866 |
Hypothetical protein, PV83 orf19 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.761 |
fig|158878.1.peg.790 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.748 |
fig|158878.1.peg.47 |
Disulfide bond regulator |
Disulphide_related_cluster |
| 0.748 |
fig|158878.1.peg.912 |
Phage holin |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| 0.744 |
fig|158878.1.peg.2020 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.737 |
fig|158878.1.peg.1999 |
hypothetical protein |
|
| 0.735 |
fig|158878.1.peg.72 |
Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1) |
Potassium_homeostasis |
| 0.733 |
fig|158878.1.peg.884 |
Integrase regulator RinA |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.724 |
fig|158878.1.peg.872 |
Hypothetical protein, PV83 orf22 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.724 |
fig|158878.1.peg.32 |
hypothetical protein |
|
| 0.722 |
fig|158878.1.peg.71 |
Osmosensitive K+ channel histidine kinase KdpD (EC 2.7.3.-) |
Potassium_homeostasis |
| 0.709 |
fig|158878.1.peg.34 |
Bleomycin resistance protein |
|
| 0.706 |
fig|158878.1.peg.76 |
hypothetical protein |
|
| 0.704 |
fig|158878.1.peg.2023 |
Hypothetical SAV0789 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.700 |
fig|158878.1.peg.69 |
Conserved hypothetical protein |
|
Connections for fig|158878.1.peg.403
| PC |
PEG |
Function |
Subsystems |
| 0.904 |
fig|158878.1.peg.410 |
hypothetical protein |
|
| 0.902 |
fig|158878.1.peg.408 |
hypothetical protein |
|
| 0.889 |
fig|158878.1.peg.400 |
lipoprotein, NLP/P60 family |
|
| 0.884 |
fig|158878.1.peg.858 |
unknown function |
|
| 0.878 |
fig|158878.1.peg.402 |
hypothetical protein |
|
| 0.872 |
fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
| 0.870 |
fig|158878.1.peg.398 |
Tetracycline resistance protein TetM |
Tetracycline_resistance,_ribosome_protection_type,Translation_elongation_factor_G_family |
| 0.863 |
fig|158878.1.peg.399 |
hypothetical protein |
|
| 0.860 |
fig|158878.1.peg.405 |
hypothetical protein |
|
| 0.858 |
fig|158878.1.peg.411 |
hypothetical protein |
|
| 0.857 |
fig|158878.1.peg.865 |
HNH homing endonuclease |
|
| 0.851 |
fig|158878.1.peg.868 |
Phage replication initiation |
|
| 0.851 |
fig|158878.1.peg.791 |
Hypothetical SAV0791 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.846 |
fig|158878.1.peg.792 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.841 |
fig|158878.1.peg.397 |
hypothetical protein |
|
| 0.838 |
fig|158878.1.peg.404 |
hypothetical protein |
|
| 0.836 |
fig|158878.1.peg.409 |
hypothetical protein |
|
| 0.836 |
fig|158878.1.peg.412 |
hypothetical protein |
|
| 0.835 |
fig|158878.1.peg.406 |
hypothetical protein |
|
| 0.832 |
fig|158878.1.peg.401 |
hypothetical protein |
|
| 0.829 |
fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| 0.824 |
fig|158878.1.peg.854 |
unknown function |
|
| 0.822 |
fig|158878.1.peg.916 |
hypothetical protein |
|
| 0.821 |
fig|158878.1.peg.1989 |
hypothetical protein within prophage phiN315 |
|
| 0.818 |
fig|158878.1.peg.395 |
hypothetical protein |
|
| 0.811 |
fig|158878.1.peg.392 |
Integrase |
|
| 0.810 |
fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| 0.796 |
fig|158878.1.peg.415 |
Transposase |
|
| 0.787 |
fig|158878.1.peg.394 |
hhypothetical protein |
|
| 0.786 |
fig|158878.1.peg.2023 |
Hypothetical SAV0789 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.775 |
fig|158878.1.peg.1999 |
hypothetical protein |
|
| 0.763 |
fig|158878.1.peg.77 |
hypothetical protein |
|
| 0.762 |
fig|158878.1.peg.790 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.761 |
fig|158878.1.peg.396 |
hypothetical protein |
|
| 0.759 |
fig|158878.1.peg.912 |
Phage holin |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| 0.748 |
fig|158878.1.peg.49 |
FIG003846: hypothetical protein |
Disulphide_related_cluster |
| 0.741 |
fig|158878.1.peg.72 |
Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1) |
Potassium_homeostasis |
| 0.740 |
fig|158878.1.peg.34 |
Bleomycin resistance protein |
|
| 0.739 |
fig|158878.1.peg.2020 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.738 |
fig|158878.1.peg.47 |
Disulfide bond regulator |
Disulphide_related_cluster |
| 0.738 |
fig|158878.1.peg.2021 |
Putative DNA helicase, superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.737 |
fig|158878.1.peg.866 |
Hypothetical protein, PV83 orf19 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.736 |
fig|158878.1.peg.58 |
hypothetical protein |
|
| 0.734 |
fig|158878.1.peg.46 |
Zn-dependent hydroxyacylglutathione hydrolase |
Disulphide_related_cluster |
| 0.734 |
fig|158878.1.peg.872 |
Hypothetical protein, PV83 orf22 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.733 |
fig|158878.1.peg.1078 |
hypothetical protein |
|
| 0.733 |
fig|158878.1.peg.32 |
hypothetical protein |
|
| 0.733 |
fig|158878.1.peg.76 |
hypothetical protein |
|
| 0.732 |
fig|158878.1.peg.69 |
Conserved hypothetical protein |
|
| 0.731 |
fig|158878.1.peg.869 |
DNA replication protein DnaC |
DNA-replication |
| 0.716 |
fig|158878.1.peg.884 |
Integrase regulator RinA |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.713 |
fig|158878.1.peg.57 |
DNA repair protein RadC truncated by transposon Tn554 |
|
| 0.711 |
fig|158878.1.peg.917 |
hypothetical protein |
|
| 0.710 |
fig|158878.1.peg.45 |
Polysulfide binding protein |
Disulphide_related_cluster |
| 0.704 |
fig|158878.1.peg.64 |
hypothetical protein |
|
Connections for fig|158878.1.peg.405
| PC |
PEG |
Function |
Subsystems |
| 0.937 |
fig|158878.1.peg.402 |
hypothetical protein |
|
| 0.925 |
fig|158878.1.peg.411 |
hypothetical protein |
|
| 0.924 |
fig|158878.1.peg.400 |
lipoprotein, NLP/P60 family |
|
| 0.913 |
fig|158878.1.peg.409 |
hypothetical protein |
|
| 0.910 |
fig|158878.1.peg.415 |
Transposase |
|
| 0.907 |
fig|158878.1.peg.408 |
hypothetical protein |
|
| 0.899 |
fig|158878.1.peg.401 |
hypothetical protein |
|
| 0.897 |
fig|158878.1.peg.412 |
hypothetical protein |
|
| 0.894 |
fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
| 0.889 |
fig|158878.1.peg.398 |
Tetracycline resistance protein TetM |
Tetracycline_resistance,_ribosome_protection_type,Translation_elongation_factor_G_family |
| 0.874 |
fig|158878.1.peg.410 |
hypothetical protein |
|
| 0.869 |
fig|158878.1.peg.792 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.862 |
fig|158878.1.peg.394 |
hhypothetical protein |
|
| 0.860 |
fig|158878.1.peg.403 |
hypothetical protein |
|
| 0.855 |
fig|158878.1.peg.399 |
hypothetical protein |
|
| 0.850 |
fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| 0.847 |
fig|158878.1.peg.865 |
HNH homing endonuclease |
|
| 0.838 |
fig|158878.1.peg.406 |
hypothetical protein |
|
| 0.838 |
fig|158878.1.peg.868 |
Phage replication initiation |
|
| 0.837 |
fig|158878.1.peg.791 |
Hypothetical SAV0791 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.831 |
fig|158878.1.peg.404 |
hypothetical protein |
|
| 0.830 |
fig|158878.1.peg.392 |
Integrase |
|
| 0.830 |
fig|158878.1.peg.397 |
hypothetical protein |
|
| 0.827 |
fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| 0.821 |
fig|158878.1.peg.395 |
hypothetical protein |
|
| 0.794 |
fig|158878.1.peg.1989 |
hypothetical protein within prophage phiN315 |
|
| 0.792 |
fig|158878.1.peg.858 |
unknown function |
|
| 0.787 |
fig|158878.1.peg.2021 |
Putative DNA helicase, superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.780 |
fig|158878.1.peg.916 |
hypothetical protein |
|
| 0.770 |
fig|158878.1.peg.869 |
DNA replication protein DnaC |
DNA-replication |
| 0.762 |
fig|158878.1.peg.866 |
Hypothetical protein, PV83 orf19 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.759 |
fig|158878.1.peg.854 |
unknown function |
|
| 0.748 |
fig|158878.1.peg.2020 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.745 |
fig|158878.1.peg.790 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.736 |
fig|158878.1.peg.884 |
Integrase regulator RinA |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.736 |
fig|158878.1.peg.912 |
Phage holin |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| 0.731 |
fig|158878.1.peg.47 |
Disulfide bond regulator |
Disulphide_related_cluster |
| 0.720 |
fig|158878.1.peg.1999 |
hypothetical protein |
|
| 0.718 |
fig|158878.1.peg.72 |
Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1) |
Potassium_homeostasis |
| 0.710 |
fig|158878.1.peg.872 |
Hypothetical protein, PV83 orf22 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.701 |
fig|158878.1.peg.71 |
Osmosensitive K+ channel histidine kinase KdpD (EC 2.7.3.-) |
Potassium_homeostasis |
Connections for fig|158878.1.peg.406
| PC |
PEG |
Function |
Subsystems |
| 0.873 |
fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| 0.868 |
fig|158878.1.peg.404 |
hypothetical protein |
|
| 0.862 |
fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| 0.862 |
fig|158878.1.peg.392 |
Integrase |
|
| 0.861 |
fig|158878.1.peg.397 |
hypothetical protein |
|
| 0.847 |
fig|158878.1.peg.411 |
hypothetical protein |
|
| 0.845 |
fig|158878.1.peg.398 |
Tetracycline resistance protein TetM |
Tetracycline_resistance,_ribosome_protection_type,Translation_elongation_factor_G_family |
| 0.842 |
fig|158878.1.peg.400 |
lipoprotein, NLP/P60 family |
|
| 0.842 |
fig|158878.1.peg.402 |
hypothetical protein |
|
| 0.840 |
fig|158878.1.peg.408 |
hypothetical protein |
|
| 0.838 |
fig|158878.1.peg.405 |
hypothetical protein |
|
| 0.835 |
fig|158878.1.peg.403 |
hypothetical protein |
|
| 0.835 |
fig|158878.1.peg.412 |
hypothetical protein |
|
| 0.830 |
fig|158878.1.peg.410 |
hypothetical protein |
|
| 0.826 |
fig|158878.1.peg.399 |
hypothetical protein |
|
| 0.825 |
fig|158878.1.peg.407 |
FIG086557: Conjugation related protein |
|
| 0.824 |
fig|158878.1.peg.415 |
Transposase |
|
| 0.816 |
fig|158878.1.peg.401 |
hypothetical protein |
|
| 0.816 |
fig|158878.1.peg.409 |
hypothetical protein |
|
| 0.805 |
fig|158878.1.peg.792 |
Hypothetical SAV0792 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.800 |
fig|158878.1.peg.791 |
Hypothetical SAV0791 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.798 |
fig|158878.1.peg.868 |
Phage replication initiation |
|
| 0.795 |
fig|158878.1.peg.395 |
hypothetical protein |
|
| 0.792 |
fig|158878.1.peg.394 |
hhypothetical protein |
|
| 0.787 |
fig|158878.1.peg.865 |
HNH homing endonuclease |
|
| 0.775 |
fig|158878.1.peg.916 |
hypothetical protein |
|
| 0.761 |
fig|158878.1.peg.858 |
unknown function |
|
| 0.760 |
fig|158878.1.peg.869 |
DNA replication protein DnaC |
DNA-replication |
| 0.758 |
fig|158878.1.peg.1989 |
hypothetical protein within prophage phiN315 |
|
| 0.744 |
fig|158878.1.peg.912 |
Phage holin |
Phage_lysis_modules,Staphylococcal_phi-Mu50B-like_prophages |
| 0.738 |
fig|158878.1.peg.854 |
unknown function |
|
| 0.729 |
fig|158878.1.peg.393 |
hypothetical protein |
|
| 0.716 |
fig|158878.1.peg.872 |
Hypothetical protein, PV83 orf22 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
| 0.714 |
fig|158878.1.peg.790 |
Hypothetical SAV0790 homolog in superantigen-encoding pathogenicity islands SaPI |
Staphylococcal_pathogenicity_islands_SaPI |
| 0.709 |
fig|158878.1.peg.866 |
Hypothetical protein, PV83 orf19 homolog [SA bacteriophages 11, Mu50B] |
Staphylococcal_phi-Mu50B-like_prophages |
It may appear tedious to go through all of these tables of connections,
but there are gems in the details. In any event, our original cluster of
hypothetical genes is now (I believe) solidly tied to a number of components
of the cellular machinery related to pathogenicity.
Pegs in Atomic Regulon 37 [ON=329 OFF=512]
This atomic regulon represents Fatty Acid Metabolism, with an additional function that converts
acetoacetyl-CoA to acetoacetate (see Synthesis and degradation of ketone bodies).
Our actual annotations for this cluster are wrong in two ways:
-
We have assigned peg.233 the function Glutaryl-CoA dehydrogenase (EC 1.3.99.7).
It should probably be Acyl coenzyme A dehydrogenase (EC 1.3.99.13).
-
peg.235, which is now labeled hypothetical protein, should be
3-oxoacid CoA-transferase (EC 2.8.3.5).
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.231 |
3-ketoacyl-CoA thiolase (EC 2.3.1.16) |
Biotin_biosynthesis,CBSS-246196.1.peg.364,Isoleucine_degradation,Polyhydroxybutyrate_metabolism,Serine-glyoxylate_cycle,n-Phenylalkanoic_acid_degradation |
| fig|158878.1.peg.232 |
Enoyl-CoA hydratase (EC 4.2.1.17) / Enoyl-CoA hydratase [valine degradation] (EC 4.2.1.17) / 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35) |
Acetyl-CoA_fermentation_to_Butyrate,Butanol_Biosynthesis,Isoleucine_degradation,Polyhydroxybutyrate_metabolism,Valine_degradation,n-Phenylalkanoic_acid_degradation |
| fig|158878.1.peg.233 |
Glutaryl-CoA dehydrogenase (EC 1.3.99.7) |
Anaerobic_benzoate_metabolism |
| fig|158878.1.peg.234 |
Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) |
Biotin_biosynthesis,n-Phenylalkanoic_acid_degradation |
| fig|158878.1.peg.235 |
hypothetical protein |
|
At least, that is how I see it.
Pegs in Atomic Regulon 42 [ON=196 OFF=641]
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.130 |
Exopolysaccharide biosynthesis glycosyltransferase EpsF (EC 2.4.1.-) |
Exopolysaccharide_Biosynthesis |
| fig|158878.1.peg.131 |
hypothetical protein |
|
| fig|158878.1.peg.2662 |
Manganese-dependent protein-tyrosine phosphatase (EC 3.1.3.48) |
Cell_envelope-associated_LytR-CpsA-Psr_transcriptional_attenuators,Exopolysaccharide_Biosynthesis |
| fig|158878.1.peg.2663 |
Tyrosine-protein kinase EpsD (EC 2.7.10.2) |
Cell_envelope-associated_LytR-CpsA-Psr_transcriptional_attenuators,Exopolysaccharide_Biosynthesis,Extracellular_Polysaccharide_Biosynthesis_of_Streptococci |
| fig|158878.1.peg.2664 |
Capsular polysaccharide biosynthesis protein capA |
|
In a state of almost total ignorance of the role of exopolysaccharides, I conjecture that both
clusters of genes relate to that function, and peg.131 in particular relates to it.
Pegs in Atomic Regulon 61 [ON=243 OFF=588]
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.329 |
Putative sugar-specific permease, SgaT/UlaA |
|
| fig|158878.1.peg.330 |
hypothetical protein |
|
| fig|158878.1.peg.331 |
PTS system, IIA component |
|
| fig|158878.1.peg.332 |
Putative transcriptional antiterminator, BglG family / PTS system, mannitol/fructose-specific IIA component (EC 2.7.1.69) |
|
I claim that clusters like this are common and terribly difficult to analyze.
Connections for fig|158878.1.peg.330
| PC |
PEG |
Function |
Subsystems |
| 0.865 |
fig|158878.1.peg.331 |
PTS system, IIA component |
|
| 0.842 |
fig|158878.1.peg.332 |
Putative transcriptional antiterminator, BglG family / PTS system, mannitol/fructose-specific IIA component (EC 2.7.1.69) |
|
| 0.826 |
fig|158878.1.peg.329 |
Putative sugar-specific permease, SgaT/UlaA |
|
| 0.733 |
fig|158878.1.peg.2641 |
PTS system, mannose-specific IIB component (EC 2.7.1.69) / PTS system, mannose-specific IIC component (EC 2.7.1.69) / PTS system, mannose-specific IIA component (EC 2.7.1.69) |
Mannose_Metabolism,Sialic_Acid_Metabolism |
Connections for fig|158878.1.peg.331
| PC |
PEG |
Function |
Subsystems |
| 0.868 |
fig|158878.1.peg.332 |
Putative transcriptional antiterminator, BglG family / PTS system, mannitol/fructose-specific IIA component (EC 2.7.1.69) |
|
| 0.865 |
fig|158878.1.peg.330 |
hypothetical protein |
|
| 0.749 |
fig|158878.1.peg.329 |
Putative sugar-specific permease, SgaT/UlaA |
|
| 0.714 |
fig|158878.1.peg.2641 |
PTS system, mannose-specific IIB component (EC 2.7.1.69) / PTS system, mannose-specific IIC component (EC 2.7.1.69) / PTS system, mannose-specific IIA component (EC 2.7.1.69) |
Mannose_Metabolism,Sialic_Acid_Metabolism |
I believe that peg.2641 and peg.2642 are co-regulated:
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.2641 |
PTS system, mannose-specific IIB component (EC 2.7.1.69) / PTS system, mannose-specific IIC component (EC 2.7.1.69) / PTS system, mannose-specific IIA component (EC 2.7.1.69) |
Mannose_Metabolism,Sialic_Acid_Metabolism |
| fig|158878.1.peg.2642 |
Mannose-6-phosphate isomerase (EC 5.3.1.8) |
Alginate_metabolism,Mannose_Metabolism |
This really does not cure our problem, but it does offer clues. In particular, it
seems like a good bet that whatever peg.2641 is used to transport is the
substrate of this PTS module. Let us examine the evidence:
- peg.331 shows a domain characterized as "PTS_IIA, PTS system, fructose/mannitol specific IIA subunit."
- peg.330 hits a domain characterized as
PTS_IIB_ascorbate: subunit IIB of enzyme II (EII) of the L-ascorbate-specific
phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS).
In this system, EII is an L-ascorbate-specific permease with two
cytoplasmic subunits (IIA and IIB) and a transmembrane channel IIC
subunit. Subunits IIA, IIB, and IIC are encoded by the sgaA, sgaB, and
sgaT genes of the E. coli sgaTBA operon. In some bacteria, the IIB
(SgaB) domain is fused C-terminal to the IIA (SgaT) domain. The IIB
domain fold includes a central four-stranded parallel open twisted
beta-sheet flanked by alpha-helices on both sides. The seven major PTS
systems with this IIB fold include ascorbate, chitobiose/lichenan,
lactose, galactitol, mannitol, fructose, and a sensory system with
similarity to the bacterial bgl system.
-
peg.329 hits a domain characterized as "ascorbate-specific PTS system enzyme IIC".
-
peg.2641 hits a domain characterized as "Phosphotransferase system, fructose-specific IIC component".
-
peg.2642 hits a domain characterized as "mannose-6-phosphate isomerase, class I".
- Finally, the SEED annotators seemed to believe that mannose was the substrate.
This "evidence" reflects the total uncertainty annotators experience when trying to guess
the substrate of a PTS module. However,
I believe that peg.330 is the IIB subunit, and the substrate is probably mannose.
The mannose-6-phosphate is converted to fructose-6-phosphate by
the mannose-6-phosphate isomerase, and the fructose-6-phosphate flows into glycolysis.
My reasoning is essentially as follows:
- The substrate of the PTS module is completely undetermined, although we can nail
down the subunits.
- The expression correlations and clustering point directly at peg.2642 as
the consumer of the substrate.
- peg.2642 appears to have the solid function Mannose-6-phosphate isomerase (EC 5.3.1.8).
- The import of mannose-6-phosphate, conversion to fructose-6-phosphate, and then catabolizing that via glycolysis is a simple model.
It would be good to check some of the conditions to verify that mannose was in the growth media.
However, we do have about 196 experiments registering ON and 641 OFF, so maybe checking a few samples
would help determine whether or not my model is accurate.
It is also worth mentioning that the PTS module might be capable of transporting both
mannose (to mannose-6-phosphate) and fructose to (fructose-6-phosphate). That would explain the
ambiguity in determining its substrate and would fit the model (more-or-less).
Pegs in Atomic Regulon 87 [ON=778 OFF=57]
Here is a strange situation. I started with a 3-gene cluster of hypotheticals.
I asked "What do these correlate with?". I stopped expanding with these.
Pearson Coefficients:
| PEG |
peg.2182 |
peg.2183 |
peg.2184 |
peg.2085 |
peg.1739 |
peg.823 |
peg.1854 |
peg.497 |
peg.372 |
peg.1752 |
peg.1839 |
peg.1214 |
peg.2382 |
peg.2368 |
peg.613 |
peg.997 |
peg.387 |
peg.1707 |
peg.682 |
peg.1881 |
peg.1625 |
peg.616 |
peg.1103 |
peg.1617 |
peg.1624 |
peg.998 |
peg.840 |
peg.1446 |
peg.2064 |
peg.1861 |
peg.1083 |
peg.1738 |
peg.1616 |
peg.1875 |
peg.1084 |
peg.704 |
peg.767 |
peg.1481 |
peg.1406 |
peg.1717 |
peg.1706 |
peg.2474 |
peg.1447 |
peg.766 |
peg.615 |
peg.2325 |
| fig|158878.1.peg.2182 |
|
0.950 |
0.939 |
0.850 |
0.874 |
0.842 |
0.866 |
0.865 |
0.837 |
0.828 |
0.811 |
0.817 |
0.780 |
0.767 |
0.797 |
0.850 |
0.755 |
0.781 |
0.782 |
0.792 |
0.807 |
0.771 |
0.765 |
0.760 |
0.774 |
0.750 |
0.845 |
0.773 |
0.754 |
0.773 |
0.780 |
0.694 |
0.766 |
0.773 |
0.778 |
0.764 |
0.730 |
0.813 |
0.755 |
0.707 |
0.755 |
0.738 |
0.772 |
0.749 |
0.760 |
0.738 |
| fig|158878.1.peg.2183 |
0.950 |
|
0.950 |
0.860 |
0.883 |
0.898 |
0.879 |
0.851 |
0.856 |
0.830 |
0.822 |
0.808 |
0.829 |
0.807 |
0.813 |
0.793 |
0.791 |
0.783 |
0.780 |
0.782 |
0.806 |
0.765 |
0.746 |
0.747 |
0.793 |
0.727 |
0.829 |
0.762 |
0.740 |
0.736 |
0.755 |
0.729 |
0.747 |
0.757 |
0.742 |
0.766 |
0.745 |
0.806 |
0.762 |
0.702 |
0.739 |
0.753 |
0.759 |
0.769 |
0.765 |
0.724 |
| fig|158878.1.peg.2184 |
0.939 |
0.950 |
|
0.862 |
0.845 |
0.883 |
0.858 |
0.841 |
0.832 |
0.778 |
0.830 |
0.818 |
0.806 |
0.780 |
0.806 |
0.779 |
0.785 |
0.739 |
0.777 |
0.752 |
0.744 |
0.702 |
0.768 |
0.738 |
0.764 |
0.699 |
0.760 |
0.722 |
0.722 |
0.705 |
0.741 |
0.664 |
0.745 |
0.752 |
0.752 |
0.742 |
0.765 |
0.816 |
0.772 |
0.683 |
0.693 |
0.702 |
0.744 |
0.800 |
0.733 |
0.733 |
| fig|158878.1.peg.2085 |
0.850 |
0.860 |
0.862 |
|
0.846 |
0.878 |
0.857 |
0.817 |
0.790 |
0.819 |
0.814 |
0.879 |
0.835 |
0.808 |
0.895 |
0.799 |
0.839 |
0.740 |
0.800 |
0.744 |
0.778 |
0.776 |
0.823 |
0.773 |
0.785 |
0.762 |
0.772 |
0.742 |
0.697 |
0.829 |
0.722 |
0.675 |
0.804 |
0.763 |
0.710 |
0.764 |
0.788 |
0.802 |
0.774 |
0.714 |
0.794 |
0.717 |
0.753 |
0.770 |
0.762 |
0.846 |
| fig|158878.1.peg.1739 |
0.874 |
0.883 |
0.845 |
0.846 |
|
0.863 |
0.915 |
0.887 |
0.818 |
0.859 |
0.862 |
0.824 |
0.832 |
0.802 |
0.820 |
0.840 |
0.756 |
0.794 |
0.851 |
0.836 |
0.818 |
0.860 |
0.817 |
0.797 |
0.773 |
0.807 |
0.789 |
0.799 |
0.784 |
0.798 |
0.822 |
0.757 |
0.820 |
0.810 |
0.810 |
0.798 |
0.735 |
0.787 |
0.785 |
0.762 |
0.777 |
0.710 |
0.783 |
0.730 |
0.748 |
0.762 |
| fig|158878.1.peg.823 |
0.842 |
0.898 |
0.883 |
0.878 |
0.863 |
|
0.864 |
0.809 |
0.830 |
0.802 |
0.824 |
0.845 |
0.891 |
0.829 |
0.854 |
0.748 |
0.853 |
0.737 |
0.775 |
0.711 |
0.748 |
0.747 |
0.752 |
0.701 |
0.774 |
0.707 |
0.715 |
0.668 |
0.677 |
0.753 |
0.650 |
0.697 |
0.738 |
0.802 |
0.642 |
0.769 |
0.807 |
0.763 |
0.784 |
0.668 |
0.700 |
0.723 |
0.703 |
0.798 |
0.777 |
0.760 |
| fig|158878.1.peg.1854 |
0.866 |
0.879 |
0.858 |
0.857 |
0.915 |
0.864 |
|
0.882 |
0.864 |
0.883 |
0.890 |
0.820 |
0.838 |
0.846 |
0.848 |
0.820 |
0.795 |
0.813 |
0.865 |
0.849 |
0.831 |
0.823 |
0.850 |
0.828 |
0.805 |
0.822 |
0.793 |
0.809 |
0.825 |
0.815 |
0.821 |
0.778 |
0.853 |
0.860 |
0.804 |
0.850 |
0.799 |
0.819 |
0.825 |
0.807 |
0.782 |
0.728 |
0.808 |
0.805 |
0.778 |
0.813 |
| fig|158878.1.peg.497 |
0.865 |
0.851 |
0.841 |
0.817 |
0.887 |
0.809 |
0.882 |
|
0.767 |
0.867 |
0.852 |
0.782 |
0.786 |
0.802 |
0.813 |
0.864 |
0.702 |
0.772 |
0.846 |
0.842 |
0.818 |
0.833 |
0.820 |
0.820 |
0.761 |
0.838 |
0.797 |
0.834 |
0.819 |
0.823 |
0.840 |
0.744 |
0.798 |
0.792 |
0.831 |
0.772 |
0.759 |
0.825 |
0.790 |
0.795 |
0.724 |
0.695 |
0.828 |
0.816 |
0.736 |
0.769 |
| fig|158878.1.peg.372 |
0.837 |
0.856 |
0.832 |
0.790 |
0.818 |
0.830 |
0.864 |
0.767 |
|
0.837 |
0.807 |
0.757 |
0.812 |
0.826 |
0.749 |
0.689 |
0.742 |
0.822 |
0.816 |
0.797 |
0.859 |
0.738 |
0.735 |
0.725 |
0.868 |
0.714 |
0.825 |
0.697 |
0.745 |
0.706 |
0.679 |
0.779 |
0.685 |
0.798 |
0.656 |
0.763 |
0.721 |
0.700 |
0.695 |
0.694 |
0.768 |
0.827 |
0.660 |
0.718 |
0.752 |
0.700 |
| fig|158878.1.peg.1752 |
0.828 |
0.830 |
0.778 |
0.819 |
0.859 |
0.802 |
0.883 |
0.867 |
0.837 |
|
0.779 |
0.742 |
0.776 |
0.860 |
0.840 |
0.792 |
0.734 |
0.812 |
0.832 |
0.863 |
0.858 |
0.885 |
0.809 |
0.814 |
0.830 |
0.842 |
0.831 |
0.840 |
0.845 |
0.817 |
0.791 |
0.811 |
0.783 |
0.760 |
0.779 |
0.763 |
0.748 |
0.762 |
0.769 |
0.827 |
0.794 |
0.781 |
0.804 |
0.749 |
0.725 |
0.799 |
| fig|158878.1.peg.1839 |
0.811 |
0.822 |
0.830 |
0.814 |
0.862 |
0.824 |
0.890 |
0.852 |
0.807 |
0.779 |
|
0.850 |
0.849 |
0.815 |
0.824 |
0.818 |
0.759 |
0.805 |
0.797 |
0.786 |
0.769 |
0.757 |
0.791 |
0.778 |
0.768 |
0.759 |
0.755 |
0.729 |
0.745 |
0.771 |
0.775 |
0.738 |
0.776 |
0.829 |
0.776 |
0.766 |
0.745 |
0.767 |
0.773 |
0.737 |
0.767 |
0.652 |
0.705 |
0.763 |
0.741 |
0.762 |
| fig|158878.1.peg.1214 |
0.817 |
0.808 |
0.818 |
0.879 |
0.824 |
0.845 |
0.820 |
0.782 |
0.757 |
0.742 |
0.850 |
|
0.862 |
0.787 |
0.865 |
0.815 |
0.834 |
0.782 |
0.720 |
0.690 |
0.718 |
0.720 |
0.712 |
0.672 |
0.717 |
0.701 |
0.743 |
0.634 |
0.597 |
0.805 |
0.636 |
0.650 |
0.712 |
0.773 |
0.637 |
0.774 |
0.755 |
0.736 |
0.713 |
0.636 |
0.776 |
0.695 |
0.662 |
0.745 |
0.780 |
0.741 |
| fig|158878.1.peg.2382 |
0.780 |
0.829 |
0.806 |
0.835 |
0.832 |
0.891 |
0.838 |
0.786 |
0.812 |
0.776 |
0.849 |
0.862 |
|
0.826 |
0.831 |
0.729 |
0.822 |
0.789 |
0.760 |
0.720 |
0.766 |
0.752 |
0.700 |
0.691 |
0.798 |
0.708 |
0.736 |
0.637 |
0.646 |
0.745 |
0.615 |
0.763 |
0.686 |
0.776 |
0.593 |
0.756 |
0.784 |
0.696 |
0.722 |
0.636 |
0.686 |
0.737 |
0.644 |
0.782 |
0.784 |
0.716 |
| fig|158878.1.peg.2368 |
0.767 |
0.807 |
0.780 |
0.808 |
0.802 |
0.829 |
0.846 |
0.802 |
0.826 |
0.860 |
0.815 |
0.787 |
0.826 |
|
0.877 |
0.758 |
0.838 |
0.827 |
0.785 |
0.757 |
0.791 |
0.792 |
0.748 |
0.773 |
0.833 |
0.788 |
0.789 |
0.748 |
0.745 |
0.792 |
0.686 |
0.860 |
0.710 |
0.725 |
0.670 |
0.774 |
0.775 |
0.747 |
0.762 |
0.786 |
0.760 |
0.765 |
0.724 |
0.784 |
0.708 |
0.769 |
| fig|158878.1.peg.613 |
0.797 |
0.813 |
0.806 |
0.895 |
0.820 |
0.854 |
0.848 |
0.813 |
0.749 |
0.840 |
0.824 |
0.865 |
0.831 |
0.877 |
|
0.813 |
0.864 |
0.770 |
0.752 |
0.714 |
0.721 |
0.765 |
0.787 |
0.739 |
0.747 |
0.772 |
0.735 |
0.714 |
0.692 |
0.833 |
0.688 |
0.700 |
0.765 |
0.740 |
0.699 |
0.761 |
0.830 |
0.764 |
0.785 |
0.750 |
0.767 |
0.687 |
0.743 |
0.808 |
0.740 |
0.829 |
| fig|158878.1.peg.997 |
0.850 |
0.793 |
0.779 |
0.799 |
0.840 |
0.748 |
0.820 |
0.864 |
0.689 |
0.792 |
0.818 |
0.815 |
0.729 |
0.758 |
0.813 |
|
0.688 |
0.741 |
0.722 |
0.746 |
0.745 |
0.792 |
0.751 |
0.791 |
0.669 |
0.807 |
0.793 |
0.760 |
0.748 |
0.845 |
0.786 |
0.656 |
0.788 |
0.758 |
0.774 |
0.690 |
0.675 |
0.754 |
0.752 |
0.745 |
0.755 |
0.595 |
0.748 |
0.681 |
0.703 |
0.737 |
| fig|158878.1.peg.387 |
0.755 |
0.791 |
0.785 |
0.839 |
0.756 |
0.853 |
0.795 |
0.702 |
0.742 |
0.734 |
0.759 |
0.834 |
0.822 |
0.838 |
0.864 |
0.688 |
|
0.730 |
0.675 |
0.605 |
0.635 |
0.650 |
0.657 |
0.628 |
0.701 |
0.636 |
0.653 |
0.597 |
0.550 |
0.743 |
0.561 |
0.686 |
0.674 |
0.705 |
0.564 |
0.785 |
0.813 |
0.735 |
0.729 |
0.626 |
0.697 |
0.671 |
0.648 |
0.787 |
0.726 |
0.735 |
| fig|158878.1.peg.1707 |
0.781 |
0.783 |
0.739 |
0.740 |
0.794 |
0.737 |
0.813 |
0.772 |
0.822 |
0.812 |
0.805 |
0.782 |
0.789 |
0.827 |
0.770 |
0.741 |
0.730 |
|
0.720 |
0.810 |
0.812 |
0.784 |
0.634 |
0.716 |
0.753 |
0.731 |
0.831 |
0.726 |
0.702 |
0.744 |
0.684 |
0.798 |
0.653 |
0.735 |
0.660 |
0.779 |
0.645 |
0.686 |
0.651 |
0.735 |
0.809 |
0.827 |
0.698 |
0.672 |
0.784 |
0.678 |
| fig|158878.1.peg.682 |
0.782 |
0.780 |
0.777 |
0.800 |
0.851 |
0.775 |
0.865 |
0.846 |
0.816 |
0.832 |
0.797 |
0.720 |
0.760 |
0.785 |
0.752 |
0.722 |
0.675 |
0.720 |
|
0.848 |
0.831 |
0.826 |
0.854 |
0.844 |
0.813 |
0.842 |
0.742 |
0.811 |
0.813 |
0.777 |
0.809 |
0.771 |
0.809 |
0.798 |
0.788 |
0.813 |
0.741 |
0.792 |
0.782 |
0.807 |
0.728 |
0.682 |
0.770 |
0.760 |
0.677 |
0.802 |
| fig|158878.1.peg.1881 |
0.792 |
0.782 |
0.752 |
0.744 |
0.836 |
0.711 |
0.849 |
0.842 |
0.797 |
0.863 |
0.786 |
0.690 |
0.720 |
0.757 |
0.714 |
0.746 |
0.605 |
0.810 |
0.848 |
|
0.869 |
0.857 |
0.786 |
0.847 |
0.779 |
0.831 |
0.807 |
0.873 |
0.863 |
0.780 |
0.847 |
0.757 |
0.777 |
0.751 |
0.810 |
0.768 |
0.617 |
0.718 |
0.700 |
0.833 |
0.770 |
0.756 |
0.811 |
0.670 |
0.704 |
0.738 |
| fig|158878.1.peg.1625 |
0.807 |
0.806 |
0.744 |
0.778 |
0.818 |
0.748 |
0.831 |
0.818 |
0.859 |
0.858 |
0.769 |
0.718 |
0.766 |
0.791 |
0.721 |
0.745 |
0.635 |
0.812 |
0.831 |
0.869 |
|
0.855 |
0.738 |
0.826 |
0.902 |
0.821 |
0.930 |
0.832 |
0.811 |
0.788 |
0.768 |
0.851 |
0.722 |
0.743 |
0.700 |
0.721 |
0.634 |
0.693 |
0.674 |
0.768 |
0.781 |
0.813 |
0.732 |
0.670 |
0.745 |
0.710 |
| fig|158878.1.peg.616 |
0.771 |
0.765 |
0.702 |
0.776 |
0.860 |
0.747 |
0.823 |
0.833 |
0.738 |
0.885 |
0.757 |
0.720 |
0.752 |
0.792 |
0.765 |
0.792 |
0.650 |
0.784 |
0.826 |
0.857 |
0.855 |
|
0.767 |
0.832 |
0.778 |
0.882 |
0.820 |
0.851 |
0.799 |
0.849 |
0.792 |
0.799 |
0.777 |
0.738 |
0.752 |
0.746 |
0.648 |
0.731 |
0.740 |
0.820 |
0.762 |
0.737 |
0.795 |
0.651 |
0.691 |
0.772 |
| fig|158878.1.peg.1103 |
0.765 |
0.746 |
0.768 |
0.823 |
0.817 |
0.752 |
0.850 |
0.820 |
0.735 |
0.809 |
0.791 |
0.712 |
0.700 |
0.748 |
0.787 |
0.751 |
0.657 |
0.634 |
0.854 |
0.786 |
0.738 |
0.767 |
|
0.860 |
0.747 |
0.845 |
0.664 |
0.805 |
0.829 |
0.781 |
0.830 |
0.661 |
0.884 |
0.754 |
0.836 |
0.738 |
0.776 |
0.779 |
0.830 |
0.790 |
0.736 |
0.596 |
0.787 |
0.755 |
0.625 |
0.854 |
| fig|158878.1.peg.1617 |
0.760 |
0.747 |
0.738 |
0.773 |
0.797 |
0.701 |
0.828 |
0.820 |
0.725 |
0.814 |
0.778 |
0.672 |
0.691 |
0.773 |
0.739 |
0.791 |
0.628 |
0.716 |
0.844 |
0.847 |
0.826 |
0.832 |
0.860 |
|
0.777 |
0.896 |
0.782 |
0.914 |
0.905 |
0.819 |
0.889 |
0.771 |
0.882 |
0.724 |
0.848 |
0.740 |
0.694 |
0.798 |
0.840 |
0.874 |
0.759 |
0.621 |
0.852 |
0.726 |
0.632 |
0.822 |
| fig|158878.1.peg.1624 |
0.774 |
0.793 |
0.764 |
0.785 |
0.773 |
0.774 |
0.805 |
0.761 |
0.868 |
0.830 |
0.768 |
0.717 |
0.798 |
0.833 |
0.747 |
0.669 |
0.701 |
0.753 |
0.813 |
0.779 |
0.902 |
0.778 |
0.747 |
0.777 |
|
0.751 |
0.841 |
0.737 |
0.755 |
0.697 |
0.681 |
0.854 |
0.671 |
0.698 |
0.641 |
0.673 |
0.708 |
0.668 |
0.690 |
0.704 |
0.718 |
0.753 |
0.644 |
0.705 |
0.661 |
0.727 |
| fig|158878.1.peg.998 |
0.750 |
0.727 |
0.699 |
0.762 |
0.807 |
0.707 |
0.822 |
0.838 |
0.714 |
0.842 |
0.759 |
0.701 |
0.708 |
0.788 |
0.772 |
0.807 |
0.636 |
0.731 |
0.842 |
0.831 |
0.821 |
0.882 |
0.845 |
0.896 |
0.751 |
|
0.776 |
0.870 |
0.859 |
0.884 |
0.815 |
0.769 |
0.847 |
0.775 |
0.792 |
0.764 |
0.708 |
0.779 |
0.822 |
0.872 |
0.767 |
0.648 |
0.827 |
0.717 |
0.690 |
0.835 |
| fig|158878.1.peg.840 |
0.845 |
0.829 |
0.760 |
0.772 |
0.789 |
0.715 |
0.793 |
0.797 |
0.825 |
0.831 |
0.755 |
0.743 |
0.736 |
0.789 |
0.735 |
0.793 |
0.653 |
0.831 |
0.742 |
0.807 |
0.930 |
0.820 |
0.664 |
0.782 |
0.841 |
0.776 |
|
0.812 |
0.768 |
0.783 |
0.737 |
0.827 |
0.656 |
0.691 |
0.678 |
0.692 |
0.614 |
0.696 |
0.653 |
0.717 |
0.810 |
0.800 |
0.706 |
0.657 |
0.741 |
0.665 |
| fig|158878.1.peg.1446 |
0.773 |
0.762 |
0.722 |
0.742 |
0.799 |
0.668 |
0.809 |
0.834 |
0.697 |
0.840 |
0.729 |
0.634 |
0.637 |
0.748 |
0.714 |
0.760 |
0.597 |
0.726 |
0.811 |
0.873 |
0.832 |
0.851 |
0.805 |
0.914 |
0.737 |
0.870 |
0.812 |
|
0.874 |
0.798 |
0.913 |
0.767 |
0.824 |
0.674 |
0.875 |
0.736 |
0.650 |
0.795 |
0.788 |
0.859 |
0.791 |
0.682 |
0.914 |
0.691 |
0.634 |
0.764 |
| fig|158878.1.peg.2064 |
0.754 |
0.740 |
0.722 |
0.697 |
0.784 |
0.677 |
0.825 |
0.819 |
0.745 |
0.845 |
0.745 |
0.597 |
0.646 |
0.745 |
0.692 |
0.748 |
0.550 |
0.702 |
0.813 |
0.863 |
0.811 |
0.799 |
0.829 |
0.905 |
0.755 |
0.859 |
0.768 |
0.874 |
|
0.755 |
0.862 |
0.761 |
0.817 |
0.744 |
0.838 |
0.709 |
0.686 |
0.727 |
0.784 |
0.862 |
0.697 |
0.616 |
0.815 |
0.721 |
0.627 |
0.756 |
| fig|158878.1.peg.1861 |
0.773 |
0.736 |
0.705 |
0.829 |
0.798 |
0.753 |
0.815 |
0.823 |
0.706 |
0.817 |
0.771 |
0.805 |
0.745 |
0.792 |
0.833 |
0.845 |
0.743 |
0.744 |
0.777 |
0.780 |
0.788 |
0.849 |
0.781 |
0.819 |
0.697 |
0.884 |
0.783 |
0.798 |
0.755 |
|
0.736 |
0.706 |
0.817 |
0.784 |
0.719 |
0.805 |
0.730 |
0.763 |
0.762 |
0.798 |
0.803 |
0.697 |
0.795 |
0.740 |
0.782 |
0.802 |
| fig|158878.1.peg.1083 |
0.780 |
0.755 |
0.741 |
0.722 |
0.822 |
0.650 |
0.821 |
0.840 |
0.679 |
0.791 |
0.775 |
0.636 |
0.615 |
0.686 |
0.688 |
0.786 |
0.561 |
0.684 |
0.809 |
0.847 |
0.768 |
0.792 |
0.830 |
0.889 |
0.681 |
0.815 |
0.737 |
0.913 |
0.862 |
0.736 |
|
0.687 |
0.850 |
0.695 |
0.953 |
0.697 |
0.629 |
0.787 |
0.778 |
0.842 |
0.743 |
0.567 |
0.850 |
0.665 |
0.579 |
0.767 |
| fig|158878.1.peg.1738 |
0.694 |
0.729 |
0.664 |
0.675 |
0.757 |
0.697 |
0.778 |
0.744 |
0.779 |
0.811 |
0.738 |
0.650 |
0.763 |
0.860 |
0.700 |
0.656 |
0.686 |
0.798 |
0.771 |
0.757 |
0.851 |
0.799 |
0.661 |
0.771 |
0.854 |
0.769 |
0.827 |
0.767 |
0.761 |
0.706 |
0.687 |
|
0.625 |
0.645 |
0.640 |
0.713 |
0.669 |
0.690 |
0.675 |
0.761 |
0.697 |
0.740 |
0.669 |
0.692 |
0.662 |
0.661 |
| fig|158878.1.peg.1616 |
0.766 |
0.747 |
0.745 |
0.804 |
0.820 |
0.738 |
0.853 |
0.798 |
0.685 |
0.783 |
0.776 |
0.712 |
0.686 |
0.710 |
0.765 |
0.788 |
0.674 |
0.653 |
0.809 |
0.777 |
0.722 |
0.777 |
0.884 |
0.882 |
0.671 |
0.847 |
0.656 |
0.824 |
0.817 |
0.817 |
0.850 |
0.625 |
|
0.768 |
0.834 |
0.751 |
0.723 |
0.804 |
0.833 |
0.822 |
0.748 |
0.551 |
0.836 |
0.715 |
0.661 |
0.849 |
| fig|158878.1.peg.1875 |
0.773 |
0.757 |
0.752 |
0.763 |
0.810 |
0.802 |
0.860 |
0.792 |
0.798 |
0.760 |
0.829 |
0.773 |
0.776 |
0.725 |
0.740 |
0.758 |
0.705 |
0.735 |
0.798 |
0.751 |
0.743 |
0.738 |
0.754 |
0.724 |
0.698 |
0.775 |
0.691 |
0.674 |
0.744 |
0.784 |
0.695 |
0.645 |
0.768 |
|
0.682 |
0.819 |
0.755 |
0.728 |
0.749 |
0.703 |
0.679 |
0.653 |
0.700 |
0.764 |
0.783 |
0.731 |
| fig|158878.1.peg.1084 |
0.778 |
0.742 |
0.752 |
0.710 |
0.810 |
0.642 |
0.804 |
0.831 |
0.656 |
0.779 |
0.776 |
0.637 |
0.593 |
0.670 |
0.699 |
0.774 |
0.564 |
0.660 |
0.788 |
0.810 |
0.700 |
0.752 |
0.836 |
0.848 |
0.641 |
0.792 |
0.678 |
0.875 |
0.838 |
0.719 |
0.953 |
0.640 |
0.834 |
0.682 |
|
0.690 |
0.660 |
0.796 |
0.797 |
0.816 |
0.715 |
0.528 |
0.835 |
0.685 |
0.566 |
0.759 |
| fig|158878.1.peg.704 |
0.764 |
0.766 |
0.742 |
0.764 |
0.798 |
0.769 |
0.850 |
0.772 |
0.763 |
0.763 |
0.766 |
0.774 |
0.756 |
0.774 |
0.761 |
0.690 |
0.785 |
0.779 |
0.813 |
0.768 |
0.721 |
0.746 |
0.738 |
0.740 |
0.673 |
0.764 |
0.692 |
0.736 |
0.709 |
0.805 |
0.697 |
0.713 |
0.751 |
0.819 |
0.690 |
|
0.778 |
0.766 |
0.745 |
0.737 |
0.736 |
0.748 |
0.772 |
0.790 |
0.785 |
0.737 |
| fig|158878.1.peg.767 |
0.730 |
0.745 |
0.765 |
0.788 |
0.735 |
0.807 |
0.799 |
0.759 |
0.721 |
0.748 |
0.745 |
0.755 |
0.784 |
0.775 |
0.830 |
0.675 |
0.813 |
0.645 |
0.741 |
0.617 |
0.634 |
0.648 |
0.776 |
0.694 |
0.708 |
0.708 |
0.614 |
0.650 |
0.686 |
0.730 |
0.629 |
0.669 |
0.723 |
0.755 |
0.660 |
0.778 |
|
0.738 |
0.815 |
0.662 |
0.625 |
0.630 |
0.703 |
0.921 |
0.701 |
0.776 |
| fig|158878.1.peg.1481 |
0.813 |
0.806 |
0.816 |
0.802 |
0.787 |
0.763 |
0.819 |
0.825 |
0.700 |
0.762 |
0.767 |
0.736 |
0.696 |
0.747 |
0.764 |
0.754 |
0.735 |
0.686 |
0.792 |
0.718 |
0.693 |
0.731 |
0.779 |
0.798 |
0.668 |
0.779 |
0.696 |
0.795 |
0.727 |
0.763 |
0.787 |
0.690 |
0.804 |
0.728 |
0.796 |
0.766 |
0.738 |
|
0.861 |
0.765 |
0.717 |
0.605 |
0.832 |
0.799 |
0.627 |
0.795 |
| fig|158878.1.peg.1406 |
0.755 |
0.762 |
0.772 |
0.774 |
0.785 |
0.784 |
0.825 |
0.790 |
0.695 |
0.769 |
0.773 |
0.713 |
0.722 |
0.762 |
0.785 |
0.752 |
0.729 |
0.651 |
0.782 |
0.700 |
0.674 |
0.740 |
0.830 |
0.840 |
0.690 |
0.822 |
0.653 |
0.788 |
0.784 |
0.762 |
0.778 |
0.675 |
0.833 |
0.749 |
0.797 |
0.745 |
0.815 |
0.861 |
|
0.789 |
0.701 |
0.558 |
0.827 |
0.816 |
0.611 |
0.837 |
| fig|158878.1.peg.1717 |
0.707 |
0.702 |
0.683 |
0.714 |
0.762 |
0.668 |
0.807 |
0.795 |
0.694 |
0.827 |
0.737 |
0.636 |
0.636 |
0.786 |
0.750 |
0.745 |
0.626 |
0.735 |
0.807 |
0.833 |
0.768 |
0.820 |
0.790 |
0.874 |
0.704 |
0.872 |
0.717 |
0.859 |
0.862 |
0.798 |
0.842 |
0.761 |
0.822 |
0.703 |
0.816 |
0.737 |
0.662 |
0.765 |
0.789 |
|
0.732 |
0.635 |
0.844 |
0.683 |
0.622 |
0.800 |
| fig|158878.1.peg.1706 |
0.755 |
0.739 |
0.693 |
0.794 |
0.777 |
0.700 |
0.782 |
0.724 |
0.768 |
0.794 |
0.767 |
0.776 |
0.686 |
0.760 |
0.767 |
0.755 |
0.697 |
0.809 |
0.728 |
0.770 |
0.781 |
0.762 |
0.736 |
0.759 |
0.718 |
0.767 |
0.810 |
0.791 |
0.697 |
0.803 |
0.743 |
0.697 |
0.748 |
0.679 |
0.715 |
0.736 |
0.625 |
0.717 |
0.701 |
0.732 |
|
0.729 |
0.730 |
0.604 |
0.706 |
0.758 |
| fig|158878.1.peg.2474 |
0.738 |
0.753 |
0.702 |
0.717 |
0.710 |
0.723 |
0.728 |
0.695 |
0.827 |
0.781 |
0.652 |
0.695 |
0.737 |
0.765 |
0.687 |
0.595 |
0.671 |
0.827 |
0.682 |
0.756 |
0.813 |
0.737 |
0.596 |
0.621 |
0.753 |
0.648 |
0.800 |
0.682 |
0.616 |
0.697 |
0.567 |
0.740 |
0.551 |
0.653 |
0.528 |
0.748 |
0.630 |
0.605 |
0.558 |
0.635 |
0.729 |
|
0.660 |
0.647 |
0.743 |
0.597 |
| fig|158878.1.peg.1447 |
0.772 |
0.759 |
0.744 |
0.753 |
0.783 |
0.703 |
0.808 |
0.828 |
0.660 |
0.804 |
0.705 |
0.662 |
0.644 |
0.724 |
0.743 |
0.748 |
0.648 |
0.698 |
0.770 |
0.811 |
0.732 |
0.795 |
0.787 |
0.852 |
0.644 |
0.827 |
0.706 |
0.914 |
0.815 |
0.795 |
0.850 |
0.669 |
0.836 |
0.700 |
0.835 |
0.772 |
0.703 |
0.832 |
0.827 |
0.844 |
0.730 |
0.660 |
|
0.740 |
0.651 |
0.771 |
| fig|158878.1.peg.766 |
0.749 |
0.769 |
0.800 |
0.770 |
0.730 |
0.798 |
0.805 |
0.816 |
0.718 |
0.749 |
0.763 |
0.745 |
0.782 |
0.784 |
0.808 |
0.681 |
0.787 |
0.672 |
0.760 |
0.670 |
0.670 |
0.651 |
0.755 |
0.726 |
0.705 |
0.717 |
0.657 |
0.691 |
0.721 |
0.740 |
0.665 |
0.692 |
0.715 |
0.764 |
0.685 |
0.790 |
0.921 |
0.799 |
0.816 |
0.683 |
0.604 |
0.647 |
0.740 |
|
0.711 |
0.748 |
| fig|158878.1.peg.615 |
0.760 |
0.765 |
0.733 |
0.762 |
0.748 |
0.777 |
0.778 |
0.736 |
0.752 |
0.725 |
0.741 |
0.780 |
0.784 |
0.708 |
0.740 |
0.703 |
0.726 |
0.784 |
0.677 |
0.704 |
0.745 |
0.691 |
0.625 |
0.632 |
0.661 |
0.690 |
0.741 |
0.634 |
0.627 |
0.782 |
0.579 |
0.662 |
0.661 |
0.783 |
0.566 |
0.785 |
0.701 |
0.627 |
0.611 |
0.622 |
0.706 |
0.743 |
0.651 |
0.711 |
|
0.652 |
| fig|158878.1.peg.2325 |
0.738 |
0.724 |
0.733 |
0.846 |
0.762 |
0.760 |
0.813 |
0.769 |
0.700 |
0.799 |
0.762 |
0.741 |
0.716 |
0.769 |
0.829 |
0.737 |
0.735 |
0.678 |
0.802 |
0.738 |
0.710 |
0.772 |
0.854 |
0.822 |
0.727 |
0.835 |
0.665 |
0.764 |
0.756 |
0.802 |
0.767 |
0.661 |
0.849 |
0.731 |
0.759 |
0.737 |
0.776 |
0.795 |
0.837 |
0.800 |
0.758 |
0.597 |
0.771 |
0.748 |
0.652 |
|
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.2182 |
alkaline shock protein 23 |
|
| fig|158878.1.peg.2183 |
Hypothetical protein SAV2183 |
|
| fig|158878.1.peg.2184 |
Hypothetical protein SAV2184 |
|
| fig|158878.1.peg.2085 |
Hypothetical protein SAV2085 |
|
| fig|158878.1.peg.1739 |
General stress protein-like protein |
|
| fig|158878.1.peg.823 |
hypothetical protein |
|
| fig|158878.1.peg.1854 |
Hypothetical protein SAV1854 |
|
| fig|158878.1.peg.497 |
YabJ, a purine regulatory protein and member of the highly conserved YjgF family |
De_Novo_Purine_Biosynthesis |
| fig|158878.1.peg.372 |
hypothetical protein |
|
| fig|158878.1.peg.1752 |
Hypothetical protein SAV1752 |
|
| fig|158878.1.peg.1839 |
Hypothetical protein SAV1839 |
At5g48545_and_At3g56490_At1g31160,CBSS-176279.3.peg.1262,EcsAB_transporter_affecting_expression_and_secretion_of_secretory_preproteins |
| fig|158878.1.peg.1214 |
hypothetical protein |
|
| fig|158878.1.peg.2382 |
General stress protein 26 |
|
| fig|158878.1.peg.2368 |
hypothetical protein similar to TpgX |
|
| fig|158878.1.peg.613 |
hypothetical protein |
|
| fig|158878.1.peg.997 |
Regulatory protein spx |
|
| fig|158878.1.peg.387 |
hypothetical protein |
|
| fig|158878.1.peg.1707 |
FIG002379: metal-dependent hydrolase |
CBSS-269801.1.peg.809 |
| fig|158878.1.peg.682 |
hypothetical protein |
|
| fig|158878.1.peg.1881 |
FIG133424: Low molecular weight protein tyrosine phosphatase (EC 3.1.3.48) |
CBSS-176280.1.peg.1561,LMPTP_YfkJ_cluster |
| fig|158878.1.peg.1625 |
sigmaB-controlled gene product |
|
| fig|158878.1.peg.616 |
Staphylococcal accessory regulator A (SarA) |
Biofilm_formation_in_Staphylococcus |
| fig|158878.1.peg.1103 |
hypothetical protein |
|
| fig|158878.1.peg.1617 |
Hypothetical protein possible functionally linked with Alanyl-tRNA synthetase |
CBSS-257314.1.peg.488 |
| fig|158878.1.peg.1624 |
hypothetical protein |
|
| fig|158878.1.peg.998 |
Negative regulator of genetic competence MecA |
|
| fig|158878.1.peg.840 |
hypothetical protein |
|
| fig|158878.1.peg.1446 |
Cell division protein GpsB, coordinates the switch between cylindrical and septal cell wall synthesis by re-localization of PBP1 |
Control_of_cell_elongation_-_division_cycle_in_Bacilli |
| fig|158878.1.peg.2064 |
RNA polymerase sigma factor SigB |
Biofilm_formation_in_Staphylococcus,Methicillin_resistance_in_Staphylococci,SigmaB_stress_responce_regulation,Transcription_initiation,_bacterial_sigma_factors |
| fig|158878.1.peg.1861 |
Peroxide stress regulator PerR, FUR family |
Oxidative_stress,Putative_hemin_transporter |
| fig|158878.1.peg.1083 |
Phosphocarrier protein of PTS system |
HPr_catabolite_repression_system,Mannitol_Utilization |
| fig|158878.1.peg.1738 |
Maebl |
|
| fig|158878.1.peg.1616 |
Putative Holliday junction resolvase (EC 3.1.-.-) |
CBSS-257314.1.peg.488,CBSS-281090.3.peg.464,CBSS-320372.3.peg.6046 |
| fig|158878.1.peg.1875 |
ThiJ/PfpI family protein |
CBSS-176280.1.peg.1561,COG2363 |
| fig|158878.1.peg.1084 |
Phosphoenolpyruvate-protein phosphotransferase of PTS system (EC 2.7.3.9) |
Mannitol_Utilization |
| fig|158878.1.peg.704 |
CsbB stress response protein |
|
| fig|158878.1.peg.767 |
FIG001886: Cytoplasmic hypothetical protein |
Cluster_containing_CofD-like_protein_and_co-occuring_with_DNA_repair |
| fig|158878.1.peg.1481 |
Elastin binding protein EbpS |
Adhesins_in_Staphylococcus |
| fig|158878.1.peg.1406 |
Tellurite resistance protein |
|
| fig|158878.1.peg.1717 |
Septation ring formation regulator EzrA |
CBSS-393130.3.peg.794,Control_of_cell_elongation_-_division_cycle_in_Bacilli |
| fig|158878.1.peg.1706 |
Universal stress protein family |
CBSS-269801.1.peg.809,Universal_stress_protein_family |
| fig|158878.1.peg.2474 |
hypothetical protein |
|
| fig|158878.1.peg.1447 |
FIG005686: hypothetical protein |
Control_of_cell_elongation_-_division_cycle_in_Bacilli |
| fig|158878.1.peg.766 |
FIG002813: LPPG:FO 2-phospho-L-lactate transferase like, CofD-like |
Cluster_containing_CofD-like_protein_and_co-occuring_with_DNA_repair |
| fig|158878.1.peg.615 |
hypothetical esterase/lipase [EC:3.1.-.-] |
|
| fig|158878.1.peg.2325 |
Hypothetical protein SAV2325 |
|
I found that I was expanding into correlations between genes that tended to always
be ON or OFF, I then wrote a simple expansion logic (which I call "broadening" a set of PEGs),
and got:
Pearson Coefficients:
| PEG |
peg.2182 |
peg.2183 |
peg.2184 |
peg.2085 |
peg.1214 |
peg.823 |
peg.1739 |
peg.1839 |
peg.372 |
peg.613 |
peg.2382 |
peg.1625 |
peg.840 |
peg.2368 |
peg.1707 |
peg.682 |
peg.387 |
peg.1624 |
peg.1881 |
peg.1875 |
peg.1861 |
peg.704 |
peg.1706 |
peg.766 |
peg.767 |
peg.615 |
peg.2325 |
peg.752 |
peg.498 |
peg.2334 |
| fig|158878.1.peg.2182 |
|
0.950 |
0.939 |
0.850 |
0.817 |
0.842 |
0.874 |
0.811 |
0.837 |
0.797 |
0.780 |
0.807 |
0.845 |
0.767 |
0.781 |
0.782 |
0.755 |
0.774 |
0.792 |
0.773 |
0.773 |
0.764 |
0.755 |
0.749 |
0.730 |
0.760 |
0.738 |
0.712 |
0.766 |
0.690 |
| fig|158878.1.peg.2183 |
0.950 |
|
0.950 |
0.860 |
0.808 |
0.898 |
0.883 |
0.822 |
0.856 |
0.813 |
0.829 |
0.806 |
0.829 |
0.807 |
0.783 |
0.780 |
0.791 |
0.793 |
0.782 |
0.757 |
0.736 |
0.766 |
0.739 |
0.769 |
0.745 |
0.765 |
0.724 |
0.715 |
0.726 |
0.717 |
| fig|158878.1.peg.2184 |
0.939 |
0.950 |
|
0.862 |
0.818 |
0.883 |
0.845 |
0.830 |
0.832 |
0.806 |
0.806 |
0.744 |
0.760 |
0.780 |
0.739 |
0.777 |
0.785 |
0.764 |
0.752 |
0.752 |
0.705 |
0.742 |
0.693 |
0.800 |
0.765 |
0.733 |
0.733 |
0.672 |
0.678 |
0.727 |
| fig|158878.1.peg.2085 |
0.850 |
0.860 |
0.862 |
|
0.879 |
0.878 |
0.846 |
0.814 |
0.790 |
0.895 |
0.835 |
0.778 |
0.772 |
0.808 |
0.740 |
0.800 |
0.839 |
0.785 |
0.744 |
0.763 |
0.829 |
0.764 |
0.794 |
0.770 |
0.788 |
0.762 |
0.846 |
0.728 |
0.713 |
0.728 |
| fig|158878.1.peg.1214 |
0.817 |
0.808 |
0.818 |
0.879 |
|
0.845 |
0.824 |
0.850 |
0.757 |
0.865 |
0.862 |
0.718 |
0.743 |
0.787 |
0.782 |
0.720 |
0.834 |
0.717 |
0.690 |
0.773 |
0.805 |
0.774 |
0.776 |
0.745 |
0.755 |
0.780 |
0.741 |
0.767 |
0.743 |
0.758 |
| fig|158878.1.peg.823 |
0.842 |
0.898 |
0.883 |
0.878 |
0.845 |
|
0.863 |
0.824 |
0.830 |
0.854 |
0.891 |
0.748 |
0.715 |
0.829 |
0.737 |
0.775 |
0.853 |
0.774 |
0.711 |
0.802 |
0.753 |
0.769 |
0.700 |
0.798 |
0.807 |
0.777 |
0.760 |
0.699 |
0.692 |
0.776 |
| fig|158878.1.peg.1739 |
0.874 |
0.883 |
0.845 |
0.846 |
0.824 |
0.863 |
|
0.862 |
0.818 |
0.820 |
0.832 |
0.818 |
0.789 |
0.802 |
0.794 |
0.851 |
0.756 |
0.773 |
0.836 |
0.810 |
0.798 |
0.798 |
0.777 |
0.730 |
0.735 |
0.748 |
0.762 |
0.718 |
0.776 |
0.692 |
| fig|158878.1.peg.1839 |
0.811 |
0.822 |
0.830 |
0.814 |
0.850 |
0.824 |
0.862 |
|
0.807 |
0.824 |
0.849 |
0.769 |
0.755 |
0.815 |
0.805 |
0.797 |
0.759 |
0.768 |
0.786 |
0.829 |
0.771 |
0.766 |
0.767 |
0.763 |
0.745 |
0.741 |
0.762 |
0.742 |
0.767 |
0.728 |
| fig|158878.1.peg.372 |
0.837 |
0.856 |
0.832 |
0.790 |
0.757 |
0.830 |
0.818 |
0.807 |
|
0.749 |
0.812 |
0.859 |
0.825 |
0.826 |
0.822 |
0.816 |
0.742 |
0.868 |
0.797 |
0.798 |
0.706 |
0.763 |
0.768 |
0.718 |
0.721 |
0.752 |
0.700 |
0.709 |
0.709 |
0.715 |
| fig|158878.1.peg.613 |
0.797 |
0.813 |
0.806 |
0.895 |
0.865 |
0.854 |
0.820 |
0.824 |
0.749 |
|
0.831 |
0.721 |
0.735 |
0.877 |
0.770 |
0.752 |
0.864 |
0.747 |
0.714 |
0.740 |
0.833 |
0.761 |
0.767 |
0.808 |
0.830 |
0.740 |
0.829 |
0.814 |
0.734 |
0.771 |
| fig|158878.1.peg.2382 |
0.780 |
0.829 |
0.806 |
0.835 |
0.862 |
0.891 |
0.832 |
0.849 |
0.812 |
0.831 |
|
0.766 |
0.736 |
0.826 |
0.789 |
0.760 |
0.822 |
0.798 |
0.720 |
0.776 |
0.745 |
0.756 |
0.686 |
0.782 |
0.784 |
0.784 |
0.716 |
0.737 |
0.739 |
0.779 |
| fig|158878.1.peg.1625 |
0.807 |
0.806 |
0.744 |
0.778 |
0.718 |
0.748 |
0.818 |
0.769 |
0.859 |
0.721 |
0.766 |
|
0.930 |
0.791 |
0.812 |
0.831 |
0.635 |
0.902 |
0.869 |
0.743 |
0.788 |
0.721 |
0.781 |
0.670 |
0.634 |
0.745 |
0.710 |
0.751 |
0.812 |
0.585 |
| fig|158878.1.peg.840 |
0.845 |
0.829 |
0.760 |
0.772 |
0.743 |
0.715 |
0.789 |
0.755 |
0.825 |
0.735 |
0.736 |
0.930 |
|
0.789 |
0.831 |
0.742 |
0.653 |
0.841 |
0.807 |
0.691 |
0.783 |
0.692 |
0.810 |
0.657 |
0.614 |
0.741 |
0.665 |
0.815 |
0.822 |
0.595 |
| fig|158878.1.peg.2368 |
0.767 |
0.807 |
0.780 |
0.808 |
0.787 |
0.829 |
0.802 |
0.815 |
0.826 |
0.877 |
0.826 |
0.791 |
0.789 |
|
0.827 |
0.785 |
0.838 |
0.833 |
0.757 |
0.725 |
0.792 |
0.774 |
0.760 |
0.784 |
0.775 |
0.708 |
0.769 |
0.842 |
0.750 |
0.776 |
| fig|158878.1.peg.1707 |
0.781 |
0.783 |
0.739 |
0.740 |
0.782 |
0.737 |
0.794 |
0.805 |
0.822 |
0.770 |
0.789 |
0.812 |
0.831 |
0.827 |
|
0.720 |
0.730 |
0.753 |
0.810 |
0.735 |
0.744 |
0.779 |
0.809 |
0.672 |
0.645 |
0.784 |
0.678 |
0.809 |
0.793 |
0.707 |
| fig|158878.1.peg.682 |
0.782 |
0.780 |
0.777 |
0.800 |
0.720 |
0.775 |
0.851 |
0.797 |
0.816 |
0.752 |
0.760 |
0.831 |
0.742 |
0.785 |
0.720 |
|
0.675 |
0.813 |
0.848 |
0.798 |
0.777 |
0.813 |
0.728 |
0.760 |
0.741 |
0.677 |
0.802 |
0.687 |
0.727 |
0.665 |
| fig|158878.1.peg.387 |
0.755 |
0.791 |
0.785 |
0.839 |
0.834 |
0.853 |
0.756 |
0.759 |
0.742 |
0.864 |
0.822 |
0.635 |
0.653 |
0.838 |
0.730 |
0.675 |
|
0.701 |
0.605 |
0.705 |
0.743 |
0.785 |
0.697 |
0.787 |
0.813 |
0.726 |
0.735 |
0.762 |
0.653 |
0.847 |
| fig|158878.1.peg.1624 |
0.774 |
0.793 |
0.764 |
0.785 |
0.717 |
0.774 |
0.773 |
0.768 |
0.868 |
0.747 |
0.798 |
0.902 |
0.841 |
0.833 |
0.753 |
0.813 |
0.701 |
|
0.779 |
0.698 |
0.697 |
0.673 |
0.718 |
0.705 |
0.708 |
0.661 |
0.727 |
0.713 |
0.708 |
0.624 |
| fig|158878.1.peg.1881 |
0.792 |
0.782 |
0.752 |
0.744 |
0.690 |
0.711 |
0.836 |
0.786 |
0.797 |
0.714 |
0.720 |
0.869 |
0.807 |
0.757 |
0.810 |
0.848 |
0.605 |
0.779 |
|
0.751 |
0.780 |
0.768 |
0.770 |
0.670 |
0.617 |
0.704 |
0.738 |
0.688 |
0.781 |
0.582 |
| fig|158878.1.peg.1875 |
0.773 |
0.757 |
0.752 |
0.763 |
0.773 |
0.802 |
0.810 |
0.829 |
0.798 |
0.740 |
0.776 |
0.743 |
0.691 |
0.725 |
0.735 |
0.798 |
0.705 |
0.698 |
0.751 |
|
0.784 |
0.819 |
0.679 |
0.764 |
0.755 |
0.783 |
0.731 |
0.655 |
0.772 |
0.767 |
| fig|158878.1.peg.1861 |
0.773 |
0.736 |
0.705 |
0.829 |
0.805 |
0.753 |
0.798 |
0.771 |
0.706 |
0.833 |
0.745 |
0.788 |
0.783 |
0.792 |
0.744 |
0.777 |
0.743 |
0.697 |
0.780 |
0.784 |
|
0.805 |
0.803 |
0.740 |
0.730 |
0.782 |
0.802 |
0.789 |
0.832 |
0.690 |
| fig|158878.1.peg.704 |
0.764 |
0.766 |
0.742 |
0.764 |
0.774 |
0.769 |
0.798 |
0.766 |
0.763 |
0.761 |
0.756 |
0.721 |
0.692 |
0.774 |
0.779 |
0.813 |
0.785 |
0.673 |
0.768 |
0.819 |
0.805 |
|
0.736 |
0.790 |
0.778 |
0.785 |
0.737 |
0.727 |
0.761 |
0.817 |
| fig|158878.1.peg.1706 |
0.755 |
0.739 |
0.693 |
0.794 |
0.776 |
0.700 |
0.777 |
0.767 |
0.768 |
0.767 |
0.686 |
0.781 |
0.810 |
0.760 |
0.809 |
0.728 |
0.697 |
0.718 |
0.770 |
0.679 |
0.803 |
0.736 |
|
0.604 |
0.625 |
0.706 |
0.758 |
0.741 |
0.694 |
0.601 |
| fig|158878.1.peg.766 |
0.749 |
0.769 |
0.800 |
0.770 |
0.745 |
0.798 |
0.730 |
0.763 |
0.718 |
0.808 |
0.782 |
0.670 |
0.657 |
0.784 |
0.672 |
0.760 |
0.787 |
0.705 |
0.670 |
0.764 |
0.740 |
0.790 |
0.604 |
|
0.921 |
0.711 |
0.748 |
0.711 |
0.714 |
0.832 |
| fig|158878.1.peg.767 |
0.730 |
0.745 |
0.765 |
0.788 |
0.755 |
0.807 |
0.735 |
0.745 |
0.721 |
0.830 |
0.784 |
0.634 |
0.614 |
0.775 |
0.645 |
0.741 |
0.813 |
0.708 |
0.617 |
0.755 |
0.730 |
0.778 |
0.625 |
0.921 |
|
0.701 |
0.776 |
0.672 |
0.662 |
0.818 |
| fig|158878.1.peg.615 |
0.760 |
0.765 |
0.733 |
0.762 |
0.780 |
0.777 |
0.748 |
0.741 |
0.752 |
0.740 |
0.784 |
0.745 |
0.741 |
0.708 |
0.784 |
0.677 |
0.726 |
0.661 |
0.704 |
0.783 |
0.782 |
0.785 |
0.706 |
0.711 |
0.701 |
|
0.652 |
0.704 |
0.789 |
0.776 |
| fig|158878.1.peg.2325 |
0.738 |
0.724 |
0.733 |
0.846 |
0.741 |
0.760 |
0.762 |
0.762 |
0.700 |
0.829 |
0.716 |
0.710 |
0.665 |
0.769 |
0.678 |
0.802 |
0.735 |
0.727 |
0.738 |
0.731 |
0.802 |
0.737 |
0.758 |
0.748 |
0.776 |
0.652 |
|
0.669 |
0.643 |
0.658 |
| fig|158878.1.peg.752 |
0.712 |
0.715 |
0.672 |
0.728 |
0.767 |
0.699 |
0.718 |
0.742 |
0.709 |
0.814 |
0.737 |
0.751 |
0.815 |
0.842 |
0.809 |
0.687 |
0.762 |
0.713 |
0.688 |
0.655 |
0.789 |
0.727 |
0.741 |
0.711 |
0.672 |
0.704 |
0.669 |
|
0.778 |
0.706 |
| fig|158878.1.peg.498 |
0.766 |
0.726 |
0.678 |
0.713 |
0.743 |
0.692 |
0.776 |
0.767 |
0.709 |
0.734 |
0.739 |
0.812 |
0.822 |
0.750 |
0.793 |
0.727 |
0.653 |
0.708 |
0.781 |
0.772 |
0.832 |
0.761 |
0.694 |
0.714 |
0.662 |
0.789 |
0.643 |
0.778 |
|
0.694 |
| fig|158878.1.peg.2334 |
0.690 |
0.717 |
0.727 |
0.728 |
0.758 |
0.776 |
0.692 |
0.728 |
0.715 |
0.771 |
0.779 |
0.585 |
0.595 |
0.776 |
0.707 |
0.665 |
0.847 |
0.624 |
0.582 |
0.767 |
0.690 |
0.817 |
0.601 |
0.832 |
0.818 |
0.776 |
0.658 |
0.706 |
0.694 |
|
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.2182 |
alkaline shock protein 23 |
|
| fig|158878.1.peg.2183 |
Hypothetical protein SAV2183 |
|
| fig|158878.1.peg.2184 |
Hypothetical protein SAV2184 |
|
| fig|158878.1.peg.2085 |
Hypothetical protein SAV2085 |
|
| fig|158878.1.peg.1214 |
hypothetical protein |
|
| fig|158878.1.peg.823 |
hypothetical protein |
|
| fig|158878.1.peg.1739 |
General stress protein-like protein |
|
| fig|158878.1.peg.1839 |
Hypothetical protein SAV1839 |
At5g48545_and_At3g56490_At1g31160,CBSS-176279.3.peg.1262,EcsAB_transporter_affecting_expression_and_secretion_of_secretory_preproteins |
| fig|158878.1.peg.372 |
hypothetical protein |
|
| fig|158878.1.peg.613 |
hypothetical protein |
|
| fig|158878.1.peg.2382 |
General stress protein 26 |
|
| fig|158878.1.peg.1625 |
sigmaB-controlled gene product |
|
| fig|158878.1.peg.840 |
hypothetical protein |
|
| fig|158878.1.peg.2368 |
hypothetical protein similar to TpgX |
|
| fig|158878.1.peg.1707 |
FIG002379: metal-dependent hydrolase |
CBSS-269801.1.peg.809 |
| fig|158878.1.peg.682 |
hypothetical protein |
|
| fig|158878.1.peg.387 |
hypothetical protein |
|
| fig|158878.1.peg.1624 |
hypothetical protein |
|
| fig|158878.1.peg.1881 |
FIG133424: Low molecular weight protein tyrosine phosphatase (EC 3.1.3.48) |
CBSS-176280.1.peg.1561,LMPTP_YfkJ_cluster |
| fig|158878.1.peg.1875 |
ThiJ/PfpI family protein |
CBSS-176280.1.peg.1561,COG2363 |
| fig|158878.1.peg.1861 |
Peroxide stress regulator PerR, FUR family |
Oxidative_stress,Putative_hemin_transporter |
| fig|158878.1.peg.704 |
CsbB stress response protein |
|
| fig|158878.1.peg.1706 |
Universal stress protein family |
CBSS-269801.1.peg.809,Universal_stress_protein_family |
| fig|158878.1.peg.766 |
FIG002813: LPPG:FO 2-phospho-L-lactate transferase like, CofD-like |
Cluster_containing_CofD-like_protein_and_co-occuring_with_DNA_repair |
| fig|158878.1.peg.767 |
FIG001886: Cytoplasmic hypothetical protein |
Cluster_containing_CofD-like_protein_and_co-occuring_with_DNA_repair |
| fig|158878.1.peg.615 |
hypothetical esterase/lipase [EC:3.1.-.-] |
|
| fig|158878.1.peg.2325 |
Hypothetical protein SAV2325 |
|
| fig|158878.1.peg.752 |
Ribosomal subunit interface protein |
Biotin_biosynthesis_Experimental,Ribosome_activity_modulation,YhgH |
| fig|158878.1.peg.498 |
Protein of unknown function identified by role in sporulation (SpoVG) |
Sporulation-associated_proteins_with_broader_functions |
| fig|158878.1.peg.2334 |
Formiminoglutamase (EC 3.5.3.8) |
Experimental_-_Histidine_Degradation,Histidine_Degradation |
These are still strange. You have a highly correlated set of essentially hypothetical
proteins.
Pegs in Atomic Regulon 94 [ON=395 OFF=385]
Let me pursue this theme a bit further. Atomic regulon 94 began as just
these three hypothetical proteins. I could find no meaningful or suggestive
hits against characterized proteins or domains.
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.1110 |
hypothetical protein |
|
| fig|158878.1.peg.1111 |
hypothetical protein |
|
| fig|158878.1.peg.1112 |
hypothetical protein |
|
Then I tried to expand the set to include just genes that that showed significant correlation scorres.
I got the following:
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.1110 |
hypothetical protein |
|
| fig|158878.1.peg.1111 |
hypothetical protein |
|
| fig|158878.1.peg.1112 |
hypothetical protein |
|
| fig|158878.1.peg.1845 |
Hypothetical protein SAV1845 |
|
| fig|158878.1.peg.1332 |
hypothetical protein |
|
| fig|158878.1.peg.1615 |
hypothetical protein |
|
| fig|158878.1.peg.1626 |
Iron-sulfur cluster regulator IscR |
At5g37530,Fe-S_cluster_assembly,Flavohaemoglobin,Iron-sulfur_cluster_assembly,Rrf2_family_transcriptional_regulators |
Again, we have a bunch of hypotheticals and (in this case) a regulator that may, or may not, be
accurately characterized.
Pegs in Atomic Regulon 97 [ON=163 OFF=655]
These seem quite innocent. When you ask for hits against characterized domains,
all three show solid hits against a domain characterized as superantigen-like protein.
If we start with this set of hypotheticals and "broaden" it,
we get
Here we see that with a modest amount of effort, one could build this into a coherent, informative
picture.
Pegs in Atomic Regulon 100 [ON=89 OFF=757]
Here is another small, innocent looking set of three genes.
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.413 |
ATP-dependent DNA helicase pcrA (EC 3.6.1.-) |
CBSS-393121.3.peg.1913,DNA_repair,_bacterial_UvrD_and_related_helicases |
| fig|158878.1.peg.414 |
FIG131328: Predicted ATP-dependent endonuclease of the OLD family |
|
| fig|158878.1.peg.415 |
Transposase |
|
When we "broaden it", we get
Pearson Coefficients:
| PEG |
peg.413 |
peg.414 |
peg.415 |
peg.399 |
peg.411 |
peg.406 |
peg.408 |
peg.392 |
peg.398 |
peg.410 |
peg.402 |
peg.397 |
peg.401 |
peg.404 |
peg.412 |
peg.395 |
peg.405 |
peg.400 |
peg.407 |
peg.403 |
peg.409 |
peg.394 |
| fig|158878.1.peg.413 |
|
0.886 |
0.814 |
0.806 |
0.829 |
0.873 |
0.836 |
0.896 |
0.841 |
0.822 |
0.829 |
0.852 |
0.805 |
0.875 |
0.810 |
0.817 |
0.827 |
0.818 |
0.814 |
0.810 |
0.788 |
0.776 |
| fig|158878.1.peg.414 |
0.886 |
|
0.853 |
0.849 |
0.850 |
0.862 |
0.845 |
0.890 |
0.841 |
0.837 |
0.849 |
0.844 |
0.816 |
0.878 |
0.841 |
0.812 |
0.850 |
0.829 |
0.809 |
0.829 |
0.825 |
0.822 |
| fig|158878.1.peg.415 |
0.814 |
0.853 |
|
0.833 |
0.917 |
0.824 |
0.864 |
0.832 |
0.842 |
0.808 |
0.905 |
0.803 |
0.890 |
0.806 |
0.907 |
0.806 |
0.910 |
0.905 |
0.838 |
0.796 |
0.927 |
0.877 |
| fig|158878.1.peg.399 |
0.806 |
0.849 |
0.833 |
|
0.855 |
0.826 |
0.880 |
0.823 |
0.840 |
0.872 |
0.878 |
0.829 |
0.841 |
0.829 |
0.843 |
0.812 |
0.855 |
0.859 |
0.820 |
0.863 |
0.835 |
0.797 |
| fig|158878.1.peg.411 |
0.829 |
0.850 |
0.917 |
0.855 |
|
0.847 |
0.898 |
0.836 |
0.881 |
0.858 |
0.925 |
0.844 |
0.904 |
0.823 |
0.911 |
0.827 |
0.925 |
0.924 |
0.876 |
0.858 |
0.920 |
0.870 |
| fig|158878.1.peg.406 |
0.873 |
0.862 |
0.824 |
0.826 |
0.847 |
|
0.840 |
0.862 |
0.845 |
0.830 |
0.842 |
0.861 |
0.816 |
0.868 |
0.835 |
0.795 |
0.838 |
0.842 |
0.825 |
0.835 |
0.816 |
0.792 |
| fig|158878.1.peg.408 |
0.836 |
0.845 |
0.864 |
0.880 |
0.898 |
0.840 |
|
0.837 |
0.886 |
0.898 |
0.924 |
0.846 |
0.892 |
0.836 |
0.877 |
0.836 |
0.907 |
0.925 |
0.898 |
0.902 |
0.886 |
0.826 |
| fig|158878.1.peg.392 |
0.896 |
0.890 |
0.832 |
0.823 |
0.836 |
0.862 |
0.837 |
|
0.839 |
0.833 |
0.832 |
0.847 |
0.821 |
0.884 |
0.837 |
0.811 |
0.830 |
0.829 |
0.816 |
0.811 |
0.806 |
0.810 |
| fig|158878.1.peg.398 |
0.841 |
0.841 |
0.842 |
0.840 |
0.881 |
0.845 |
0.886 |
0.839 |
|
0.870 |
0.895 |
0.864 |
0.860 |
0.838 |
0.847 |
0.800 |
0.889 |
0.885 |
0.871 |
0.870 |
0.852 |
0.819 |
| fig|158878.1.peg.410 |
0.822 |
0.837 |
0.808 |
0.872 |
0.858 |
0.830 |
0.898 |
0.833 |
0.870 |
|
0.878 |
0.843 |
0.839 |
0.842 |
0.838 |
0.827 |
0.874 |
0.884 |
0.869 |
0.904 |
0.841 |
0.804 |
| fig|158878.1.peg.402 |
0.829 |
0.849 |
0.905 |
0.878 |
0.925 |
0.842 |
0.924 |
0.832 |
0.895 |
0.878 |
|
0.840 |
0.911 |
0.827 |
0.903 |
0.824 |
0.937 |
0.932 |
0.903 |
0.878 |
0.914 |
0.869 |
| fig|158878.1.peg.397 |
0.852 |
0.844 |
0.803 |
0.829 |
0.844 |
0.861 |
0.846 |
0.847 |
0.864 |
0.843 |
0.840 |
|
0.815 |
0.850 |
0.829 |
0.807 |
0.830 |
0.843 |
0.814 |
0.841 |
0.809 |
0.790 |
| fig|158878.1.peg.401 |
0.805 |
0.816 |
0.890 |
0.841 |
0.904 |
0.816 |
0.892 |
0.821 |
0.860 |
0.839 |
0.911 |
0.815 |
|
0.787 |
0.900 |
0.778 |
0.899 |
0.913 |
0.870 |
0.832 |
0.913 |
0.847 |
| fig|158878.1.peg.404 |
0.875 |
0.878 |
0.806 |
0.829 |
0.823 |
0.868 |
0.836 |
0.884 |
0.838 |
0.842 |
0.827 |
0.850 |
0.787 |
|
0.814 |
0.816 |
0.831 |
0.821 |
0.808 |
0.838 |
0.787 |
0.802 |
| fig|158878.1.peg.412 |
0.810 |
0.841 |
0.907 |
0.843 |
0.911 |
0.835 |
0.877 |
0.837 |
0.847 |
0.838 |
0.903 |
0.829 |
0.900 |
0.814 |
|
0.807 |
0.897 |
0.902 |
0.850 |
0.836 |
0.915 |
0.885 |
| fig|158878.1.peg.395 |
0.817 |
0.812 |
0.806 |
0.812 |
0.827 |
0.795 |
0.836 |
0.811 |
0.800 |
0.827 |
0.824 |
0.807 |
0.778 |
0.816 |
0.807 |
|
0.821 |
0.823 |
0.795 |
0.818 |
0.797 |
0.797 |
| fig|158878.1.peg.405 |
0.827 |
0.850 |
0.910 |
0.855 |
0.925 |
0.838 |
0.907 |
0.830 |
0.889 |
0.874 |
0.937 |
0.830 |
0.899 |
0.831 |
0.897 |
0.821 |
|
0.924 |
0.894 |
0.860 |
0.913 |
0.862 |
| fig|158878.1.peg.400 |
0.818 |
0.829 |
0.905 |
0.859 |
0.924 |
0.842 |
0.925 |
0.829 |
0.885 |
0.884 |
0.932 |
0.843 |
0.913 |
0.821 |
0.902 |
0.823 |
0.924 |
|
0.901 |
0.889 |
0.925 |
0.858 |
| fig|158878.1.peg.407 |
0.814 |
0.809 |
0.838 |
0.820 |
0.876 |
0.825 |
0.898 |
0.816 |
0.871 |
0.869 |
0.903 |
0.814 |
0.870 |
0.808 |
0.850 |
0.795 |
0.894 |
0.901 |
|
0.872 |
0.865 |
0.823 |
| fig|158878.1.peg.403 |
0.810 |
0.829 |
0.796 |
0.863 |
0.858 |
0.835 |
0.902 |
0.811 |
0.870 |
0.904 |
0.878 |
0.841 |
0.832 |
0.838 |
0.836 |
0.818 |
0.860 |
0.889 |
0.872 |
|
0.836 |
0.787 |
| fig|158878.1.peg.409 |
0.788 |
0.825 |
0.927 |
0.835 |
0.920 |
0.816 |
0.886 |
0.806 |
0.852 |
0.841 |
0.914 |
0.809 |
0.913 |
0.787 |
0.915 |
0.797 |
0.913 |
0.925 |
0.865 |
0.836 |
|
0.883 |
| fig|158878.1.peg.394 |
0.776 |
0.822 |
0.877 |
0.797 |
0.870 |
0.792 |
0.826 |
0.810 |
0.819 |
0.804 |
0.869 |
0.790 |
0.847 |
0.802 |
0.885 |
0.797 |
0.862 |
0.858 |
0.823 |
0.787 |
0.883 |
|
What is revealed is a pretty large cluster of genes that probably play a role in
pathogenicity. A skilled annotator would spot this without needing the expression data,
but with the expression data a mechanical protocol can easily lay these clusters out to
be consistently annotated.
Pegs in Atomic Regulon 102 [ON=197 OFF=643]
Here again we have a set of three seemingly isolated genes. When we "broaden" the set, we get
Here, we link two groups of genes, both associated with virulence (see Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection for a discussion). Most importantly, the link just falls out.
No skill or insight was needed).
Pegs in Atomic Regulon 103 [ON=222 OFF=610]
Here is a simple example. We have three genes that all seem to be poorly characterized.
Functions in Staphylococcus aureus subsp. aureus Mu50
| PEG |
Function |
Subsystems |
| fig|158878.1.peg.2468 |
FIG018700: hypothetical protein |
CBSS-208964.1.peg.4832 |
| fig|158878.1.peg.2469 |
FIG016157: Similar to nicotianamine synthase |
CBSS-208964.1.peg.4832 |
| fig|158878.1.peg.2470 |
FIG016473: Diaminopimelate epimerase homolog |
CBSS-208964.1.peg.4832 |
When we "broaden" the set, we get
Here we see that we should be looking at a single big cluster from peg.2464 to peg.2470.
The cluster encodes a 5-gene transport along with 3 genes that do something associated. Is that it?
Well, maybe not. If you take the transport genes and look for domains, what you find is
that the substrate-binding gene (peg.2467 has a very strong hit against a a domain
that is described as
PBP2_NikA : The substrate-binding component of an ABC-type nickel import system contains
the type 2 periplasmic binding fold
The substrate-binding component of an ABC-type nickel import system
contains the type 2 periplasmic binding fold. This family represents
the periplasmic substrate-binding domain of nickel transport system,
which functions in the import of nickel and in the control of
chemotactic response away from nickel. The ATP-binding cassette (ABC)
type nickel transport system is comprised of five subunits NikABCDE:
the two pore-forming integral inner membrane proteins NikB and NikC;
the two inner membrane-associated proteins with ATPase activity NikD
and NikE; and the periplasmic nickel binding NikA, the initial nickel
receptor. The oligopeptide-binding protein OppA and the
dipeptide-binding protein DppA show significant sequence similarity to
NikA. The DppA binds dipeptides and some tripeptides and is involved
in chemotaxis toward dipeptides, whereas the OppA binds peptides of a
wide range of lengths (2-35 amino acid residues) and plays a role in
recycling of cell wall peptides, which precludes any involvement in
chemotaxis. Most of other periplasmic binding proteins are comprised
of only two globular subdomains corresponding to domains I and III of
the dipeptide/oligopeptide binding proteins. The structural topology
of these domains is most similar to that of the type 2 periplasmic
binding proteins (PBP2), which are responsible for the uptake of a
variety of substrates such as phosphate, sulfate, polysaccharides,
lysine/arginine/ornithine, and histidine. The PBP2 bind their ligand
in the cleft between these domains in a manner resembling a Venus
flytrap. After binding their specific ligand with high affinity, they
can interact with a cognate membrane transport complex comprised of
two integral membrane domains and two cytoplasmically located ATPase
domains. This interaction triggers the ligand translocation across the
cytoplasmic membrane energized by ATP hydrolysis. Besides transport
proteins, the PBP2 superfamily includes the ligand-binding domains
from ionotropic glutamate receptors, LysR-type transcriptional
regulators, and unorthodox sensor proteins involved in signal
transduction.
If we look at
A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection
we see
Summary
The oligopeptide transport systems Opp belong to the
nickel/peptide/opine PepT subfamily of ABC-transporters. The
opportunist pathogen Staphylococcus aureus encodes four putative Opps
and one orphean substrate binding protein Opp5A. Here, we report that
the Opp2 permease complex (Opp2BCDF) and Opp5A are involved in nickel
uptake and then renamed them NikBCDE and NikA respectively. S. aureus
carries also a high-affinity nickel transporter NixA belonging to the
NiCoT family of secondary transporters. The activity of these two
nickel transporters determine that of urease, a multimeric
nickel-dependent enzyme mainly involved in the neutralization of
acidic environments. However, only the Nik system was responsible for
the neutralization and deposit of pH-dependent crystals in human
urine. Inactivation of the nik genes affected bacterial colonization
of mouse urinary tract, as well as the 50% infective dose levels
compared with the parental and nixA strains. Finally, complementation
of the nik mutations restored bacterial colonization. Together, our
results suggest a role for the Nik system in the urinary tract
infection by S. aureus, probably due to the urease-mediated pH
increase of the urine.
Note that what I am saying is that the three nontransport genes may be implicated
in a nickel-related activity.
Someone who understands the role of nickel in the cell needs to look at this.
Pegs in Atomic Regulon 105 [ON=107 OFF=738]
Here, again, we have what looks like a small cluster of hypotheticals.
But, when we broaden it we get
Pearson Coefficients:
| PEG |
peg.75 |
peg.76 |
peg.77 |
peg.72 |
peg.69 |
peg.49 |
peg.47 |
peg.57 |
peg.74 |
peg.46 |
peg.44 |
peg.71 |
peg.80 |
peg.70 |
peg.34 |
peg.81 |
peg.58 |
peg.45 |
peg.31 |
peg.35 |
peg.64 |
peg.60 |
peg.67 |
peg.73 |
peg.33 |
peg.68 |
peg.63 |
peg.32 |
peg.866 |
| fig|158878.1.peg.75 |
|
0.901 |
0.777 |
0.901 |
0.886 |
0.802 |
0.824 |
0.817 |
0.861 |
0.803 |
0.811 |
0.879 |
0.867 |
0.889 |
0.767 |
0.872 |
0.791 |
0.798 |
0.769 |
0.758 |
0.742 |
0.746 |
0.791 |
0.765 |
0.725 |
0.766 |
0.693 |
0.684 |
0.723 |
| fig|158878.1.peg.76 |
0.901 |
|
0.832 |
0.929 |
0.912 |
0.854 |
0.864 |
0.854 |
0.880 |
0.844 |
0.833 |
0.898 |
0.869 |
0.882 |
0.817 |
0.863 |
0.823 |
0.838 |
0.791 |
0.789 |
0.782 |
0.776 |
0.790 |
0.768 |
0.753 |
0.756 |
0.728 |
0.760 |
0.764 |
| fig|158878.1.peg.77 |
0.777 |
0.832 |
|
0.807 |
0.829 |
0.832 |
0.773 |
0.797 |
0.786 |
0.792 |
0.778 |
0.754 |
0.798 |
0.753 |
0.804 |
0.734 |
0.791 |
0.781 |
0.705 |
0.727 |
0.755 |
0.806 |
0.693 |
0.606 |
0.790 |
0.639 |
0.735 |
0.773 |
0.667 |
| fig|158878.1.peg.72 |
0.901 |
0.929 |
0.807 |
|
0.922 |
0.850 |
0.878 |
0.850 |
0.879 |
0.862 |
0.839 |
0.925 |
0.867 |
0.871 |
0.827 |
0.871 |
0.835 |
0.838 |
0.806 |
0.815 |
0.805 |
0.754 |
0.778 |
0.799 |
0.740 |
0.769 |
0.738 |
0.740 |
0.773 |
| fig|158878.1.peg.69 |
0.886 |
0.912 |
0.829 |
0.922 |
|
0.844 |
0.857 |
0.826 |
0.857 |
0.844 |
0.814 |
0.890 |
0.869 |
0.870 |
0.823 |
0.864 |
0.810 |
0.815 |
0.770 |
0.790 |
0.795 |
0.746 |
0.778 |
0.760 |
0.741 |
0.756 |
0.728 |
0.740 |
0.746 |
| fig|158878.1.peg.49 |
0.802 |
0.854 |
0.832 |
0.850 |
0.844 |
|
0.863 |
0.849 |
0.818 |
0.875 |
0.857 |
0.821 |
0.832 |
0.793 |
0.905 |
0.796 |
0.830 |
0.863 |
0.834 |
0.850 |
0.810 |
0.810 |
0.720 |
0.647 |
0.793 |
0.684 |
0.787 |
0.765 |
0.685 |
| fig|158878.1.peg.47 |
0.824 |
0.864 |
0.773 |
0.878 |
0.857 |
0.863 |
|
0.892 |
0.848 |
0.877 |
0.869 |
0.857 |
0.787 |
0.796 |
0.836 |
0.812 |
0.861 |
0.864 |
0.847 |
0.836 |
0.826 |
0.789 |
0.748 |
0.774 |
0.778 |
0.769 |
0.759 |
0.771 |
0.746 |
| fig|158878.1.peg.57 |
0.817 |
0.854 |
0.797 |
0.850 |
0.826 |
0.849 |
0.892 |
|
0.826 |
0.865 |
0.866 |
0.802 |
0.805 |
0.778 |
0.793 |
0.777 |
0.869 |
0.881 |
0.808 |
0.772 |
0.812 |
0.838 |
0.751 |
0.680 |
0.795 |
0.749 |
0.785 |
0.737 |
0.702 |
| fig|158878.1.peg.74 |
0.861 |
0.880 |
0.786 |
0.879 |
0.857 |
0.818 |
0.848 |
0.826 |
|
0.802 |
0.824 |
0.859 |
0.831 |
0.878 |
0.790 |
0.858 |
0.785 |
0.819 |
0.815 |
0.804 |
0.748 |
0.745 |
0.797 |
0.799 |
0.762 |
0.816 |
0.691 |
0.684 |
0.752 |
| fig|158878.1.peg.46 |
0.803 |
0.844 |
0.792 |
0.862 |
0.844 |
0.875 |
0.877 |
0.865 |
0.802 |
|
0.853 |
0.821 |
0.807 |
0.782 |
0.828 |
0.793 |
0.841 |
0.832 |
0.795 |
0.804 |
0.816 |
0.794 |
0.723 |
0.669 |
0.763 |
0.706 |
0.772 |
0.740 |
0.696 |
| fig|158878.1.peg.44 |
0.811 |
0.833 |
0.778 |
0.839 |
0.814 |
0.857 |
0.869 |
0.866 |
0.824 |
0.853 |
|
0.806 |
0.806 |
0.783 |
0.826 |
0.806 |
0.829 |
0.875 |
0.839 |
0.819 |
0.810 |
0.794 |
0.751 |
0.697 |
0.784 |
0.720 |
0.758 |
0.699 |
0.700 |
| fig|158878.1.peg.71 |
0.879 |
0.898 |
0.754 |
0.925 |
0.890 |
0.821 |
0.857 |
0.802 |
0.859 |
0.821 |
0.806 |
|
0.834 |
0.871 |
0.830 |
0.903 |
0.769 |
0.793 |
0.835 |
0.846 |
0.754 |
0.662 |
0.749 |
0.846 |
0.669 |
0.747 |
0.666 |
0.696 |
0.757 |
| fig|158878.1.peg.80 |
0.867 |
0.869 |
0.798 |
0.867 |
0.869 |
0.832 |
0.787 |
0.805 |
0.831 |
0.807 |
0.806 |
0.834 |
|
0.844 |
0.780 |
0.848 |
0.794 |
0.806 |
0.743 |
0.723 |
0.748 |
0.792 |
0.788 |
0.683 |
0.741 |
0.728 |
0.714 |
0.660 |
0.674 |
| fig|158878.1.peg.70 |
0.889 |
0.882 |
0.753 |
0.871 |
0.870 |
0.793 |
0.796 |
0.778 |
0.878 |
0.782 |
0.783 |
0.871 |
0.844 |
|
0.779 |
0.870 |
0.742 |
0.763 |
0.806 |
0.794 |
0.701 |
0.698 |
0.776 |
0.770 |
0.706 |
0.762 |
0.652 |
0.639 |
0.688 |
| fig|158878.1.peg.34 |
0.767 |
0.817 |
0.804 |
0.827 |
0.823 |
0.905 |
0.836 |
0.793 |
0.790 |
0.828 |
0.826 |
0.830 |
0.780 |
0.779 |
|
0.795 |
0.784 |
0.816 |
0.880 |
0.929 |
0.789 |
0.731 |
0.659 |
0.661 |
0.757 |
0.638 |
0.737 |
0.750 |
0.661 |
| fig|158878.1.peg.81 |
0.872 |
0.863 |
0.734 |
0.871 |
0.864 |
0.796 |
0.812 |
0.777 |
0.858 |
0.793 |
0.806 |
0.903 |
0.848 |
0.870 |
0.795 |
|
0.733 |
0.775 |
0.816 |
0.810 |
0.723 |
0.657 |
0.782 |
0.784 |
0.654 |
0.772 |
0.651 |
0.569 |
0.737 |
| fig|158878.1.peg.58 |
0.791 |
0.823 |
0.791 |
0.835 |
0.810 |
0.830 |
0.861 |
0.869 |
0.785 |
0.841 |
0.829 |
0.769 |
0.794 |
0.742 |
0.784 |
0.733 |
|
0.840 |
0.753 |
0.746 |
0.845 |
0.880 |
0.727 |
0.646 |
0.799 |
0.706 |
0.826 |
0.778 |
0.739 |
| fig|158878.1.peg.45 |
0.798 |
0.838 |
0.781 |
0.838 |
0.815 |
0.863 |
0.864 |
0.881 |
0.819 |
0.832 |
0.875 |
0.793 |
0.806 |
0.763 |
0.816 |
0.775 |
0.840 |
|
0.822 |
0.777 |
0.802 |
0.819 |
0.756 |
0.697 |
0.781 |
0.730 |
0.769 |
0.724 |
0.708 |
| fig|158878.1.peg.31 |
0.769 |
0.791 |
0.705 |
0.806 |
0.770 |
0.834 |
0.847 |
0.808 |
0.815 |
0.795 |
0.839 |
0.835 |
0.743 |
0.806 |
0.880 |
0.816 |
0.753 |
0.822 |
|
0.928 |
0.740 |
0.679 |
0.700 |
0.773 |
0.727 |
0.696 |
0.674 |
0.650 |
0.644 |
| fig|158878.1.peg.35 |
0.758 |
0.789 |
0.727 |
0.815 |
0.790 |
0.850 |
0.836 |
0.772 |
0.804 |
0.804 |
0.819 |
0.846 |
0.723 |
0.794 |
0.929 |
0.810 |
0.746 |
0.777 |
0.928 |
|
0.746 |
0.649 |
0.659 |
0.748 |
0.723 |
0.659 |
0.671 |
0.689 |
0.662 |
| fig|158878.1.peg.64 |
0.742 |
0.782 |
0.755 |
0.805 |
0.795 |
0.810 |
0.826 |
0.812 |
0.748 |
0.816 |
0.810 |
0.754 |
0.748 |
0.701 |
0.789 |
0.723 |
0.845 |
0.802 |
0.740 |
0.746 |
|
0.774 |
0.660 |
0.610 |
0.737 |
0.660 |
0.845 |
0.724 |
0.707 |
| fig|158878.1.peg.60 |
0.746 |
0.776 |
0.806 |
0.754 |
0.746 |
0.810 |
0.789 |
0.838 |
0.745 |
0.794 |
0.794 |
0.662 |
0.792 |
0.698 |
0.731 |
0.657 |
0.880 |
0.819 |
0.679 |
0.649 |
0.774 |
|
0.703 |
0.503 |
0.827 |
0.658 |
0.809 |
0.743 |
0.619 |
| fig|158878.1.peg.67 |
0.791 |
0.790 |
0.693 |
0.778 |
0.778 |
0.720 |
0.748 |
0.751 |
0.797 |
0.723 |
0.751 |
0.749 |
0.788 |
0.776 |
0.659 |
0.782 |
0.727 |
0.756 |
0.700 |
0.659 |
0.660 |
0.703 |
|
0.690 |
0.663 |
0.819 |
0.680 |
0.584 |
0.663 |
| fig|158878.1.peg.73 |
0.765 |
0.768 |
0.606 |
0.799 |
0.760 |
0.647 |
0.774 |
0.680 |
0.799 |
0.669 |
0.697 |
0.846 |
0.683 |
0.770 |
0.661 |
0.784 |
0.646 |
0.697 |
0.773 |
0.748 |
0.610 |
0.503 |
0.690 |
|
0.563 |
0.729 |
0.479 |
0.590 |
0.719 |
| fig|158878.1.peg.33 |
0.725 |
0.753 |
0.790 |
0.740 |
0.741 |
0.793 |
0.778 |
0.795 |
0.762 |
0.763 |
0.784 |
0.669 |
0.741 |
0.706 |
0.757 |
0.654 |
0.799 |
0.781 |
0.727 |
0.723 |
0.737 |
0.827 |
0.663 |
0.563 |
|
0.660 |
0.734 |
0.767 |
0.596 |
| fig|158878.1.peg.68 |
0.766 |
0.756 |
0.639 |
0.769 |
0.756 |
0.684 |
0.769 |
0.749 |
0.816 |
0.706 |
0.720 |
0.747 |
0.728 |
0.762 |
0.638 |
0.772 |
0.706 |
0.730 |
0.696 |
0.659 |
0.660 |
0.658 |
0.819 |
0.729 |
0.660 |
|
0.654 |
0.567 |
0.729 |
| fig|158878.1.peg.63 |
0.693 |
0.728 |
0.735 |
0.738 |
0.728 |
0.787 |
0.759 |
0.785 |
0.691 |
0.772 |
0.758 |
0.666 |
0.714 |
0.652 |
0.737 |
0.651 |
0.826 |
0.769 |
0.674 |
0.671 |
0.845 |
0.809 |
0.680 |
0.479 |
0.734 |
0.654 |
|
0.670 |
0.612 |
| fig|158878.1.peg.32 |
0.684 |
0.760 |
0.773 |
0.740 |
0.740 |
0.765 |
0.771 |
0.737 |
0.684 |
0.740 |
0.699 |
0.696 |
0.660 |
0.639 |
0.750 |
0.569 |
0.778 |
0.724 |
0.650 |
0.689 |
0.724 |
0.743 |
0.584 |
0.590 |
0.767 |
0.567 |
0.670 |
|
0.587 |
| fig|158878.1.peg.866 |
0.723 |
0.764 |
0.667 |
0.773 |
0.746 |
0.685 |
0.746 |
0.702 |
0.752 |
0.696 |
0.700 |
0.757 |
0.674 |
0.688 |
0.661 |
0.737 |
0.739 |
0.708 |
0.644 |
0.662 |
0.707 |
0.619 |
0.663 |
0.719 |
0.596 |
0.729 |
0.612 |
0.587 |
|
Again, we see a glimpse of machinery related to stress, and perhaps playing a role in interaction with the host.
Pegs in Atomic Regulon 109 [ON=177 OFF=651]
This is again a small group that can easily be broadened into a larger cluster
that more accurately depicts a prophage:
Here is the larger cluster:
Switching to Shewanella for a while
Here is one from Shewanella. These genes are ON in 99 of 238 experiments that we have for Shewanells:
Pearson Coefficients: Get All Related PEGs
| PEG |
peg.2690 |
peg.2691 |
peg.2692 |
peg.2693 |
peg.2694 |
peg.2695 |
peg.2696 |
peg.2697 |
peg.2698 |
peg.2699 |
peg.2700 |
peg.2701 |
peg.2702 |
peg.2682 |
peg.2680 |
peg.2683 |
peg.2681 |
peg.2675 |
peg.2689 |
peg.2684 |
peg.2707 |
peg.2677 |
peg.2676 |
peg.2688 |
peg.2679 |
peg.2704 |
peg.2705 |
peg.2706 |
| fig|211586.9.peg.2690 |
|
0.895 |
0.905 |
0.865 |
0.909 |
0.904 |
0.845 |
0.838 |
0.840 |
0.873 |
0.811 |
0.748 |
0.713 |
0.828 |
0.859 |
0.819 |
0.852 |
0.735 |
0.865 |
0.835 |
0.680 |
0.754 |
0.746 |
0.735 |
0.715 |
0.705 |
0.667 |
0.703 |
| fig|211586.9.peg.2691 |
0.895 |
|
0.966 |
0.950 |
0.927 |
0.928 |
0.894 |
0.906 |
0.868 |
0.888 |
0.928 |
0.833 |
0.744 |
0.845 |
0.856 |
0.850 |
0.865 |
0.828 |
0.826 |
0.738 |
0.709 |
0.778 |
0.760 |
0.709 |
0.776 |
0.623 |
0.639 |
0.650 |
| fig|211586.9.peg.2692 |
0.905 |
0.966 |
|
0.955 |
0.936 |
0.949 |
0.904 |
0.918 |
0.895 |
0.902 |
0.924 |
0.880 |
0.788 |
0.874 |
0.862 |
0.860 |
0.894 |
0.820 |
0.845 |
0.776 |
0.765 |
0.768 |
0.768 |
0.737 |
0.763 |
0.652 |
0.666 |
0.702 |
| fig|211586.9.peg.2693 |
0.865 |
0.950 |
0.955 |
|
0.939 |
0.956 |
0.920 |
0.941 |
0.917 |
0.910 |
0.945 |
0.907 |
0.812 |
0.906 |
0.872 |
0.892 |
0.904 |
0.841 |
0.857 |
0.801 |
0.818 |
0.767 |
0.771 |
0.786 |
0.770 |
0.694 |
0.732 |
0.711 |
| fig|211586.9.peg.2694 |
0.909 |
0.927 |
0.936 |
0.939 |
|
0.979 |
0.959 |
0.969 |
0.930 |
0.944 |
0.886 |
0.859 |
0.742 |
0.916 |
0.863 |
0.898 |
0.883 |
0.850 |
0.926 |
0.859 |
0.812 |
0.775 |
0.739 |
0.816 |
0.732 |
0.775 |
0.811 |
0.798 |
| fig|211586.9.peg.2695 |
0.904 |
0.928 |
0.949 |
0.956 |
0.979 |
|
0.950 |
0.967 |
0.926 |
0.935 |
0.900 |
0.879 |
0.769 |
0.929 |
0.877 |
0.897 |
0.895 |
0.840 |
0.905 |
0.853 |
0.828 |
0.775 |
0.743 |
0.817 |
0.729 |
0.768 |
0.804 |
0.788 |
| fig|211586.9.peg.2696 |
0.845 |
0.894 |
0.904 |
0.920 |
0.959 |
0.950 |
|
0.971 |
0.957 |
0.953 |
0.892 |
0.903 |
0.780 |
0.908 |
0.856 |
0.917 |
0.884 |
0.901 |
0.907 |
0.831 |
0.814 |
0.804 |
0.767 |
0.812 |
0.778 |
0.799 |
0.849 |
0.809 |
| fig|211586.9.peg.2697 |
0.838 |
0.906 |
0.918 |
0.941 |
0.969 |
0.967 |
0.971 |
|
0.945 |
0.949 |
0.910 |
0.904 |
0.774 |
0.929 |
0.850 |
0.913 |
0.885 |
0.887 |
0.909 |
0.844 |
0.850 |
0.792 |
0.748 |
0.829 |
0.754 |
0.784 |
0.833 |
0.802 |
| fig|211586.9.peg.2698 |
0.840 |
0.868 |
0.895 |
0.917 |
0.930 |
0.926 |
0.957 |
0.945 |
|
0.963 |
0.893 |
0.917 |
0.811 |
0.916 |
0.876 |
0.924 |
0.915 |
0.886 |
0.908 |
0.862 |
0.822 |
0.842 |
0.820 |
0.821 |
0.821 |
0.822 |
0.846 |
0.817 |
| fig|211586.9.peg.2699 |
0.873 |
0.888 |
0.902 |
0.910 |
0.944 |
0.935 |
0.953 |
0.949 |
0.963 |
|
0.897 |
0.891 |
0.809 |
0.892 |
0.867 |
0.894 |
0.896 |
0.866 |
0.895 |
0.859 |
0.822 |
0.851 |
0.798 |
0.797 |
0.812 |
0.821 |
0.830 |
0.834 |
| fig|211586.9.peg.2700 |
0.811 |
0.928 |
0.924 |
0.945 |
0.886 |
0.900 |
0.892 |
0.910 |
0.893 |
0.897 |
|
0.918 |
0.837 |
0.844 |
0.835 |
0.825 |
0.864 |
0.818 |
0.777 |
0.706 |
0.782 |
0.780 |
0.766 |
0.670 |
0.795 |
0.658 |
0.691 |
0.653 |
| fig|211586.9.peg.2701 |
0.748 |
0.833 |
0.880 |
0.907 |
0.859 |
0.879 |
0.903 |
0.904 |
0.917 |
0.891 |
0.918 |
|
0.858 |
0.871 |
0.815 |
0.870 |
0.875 |
0.833 |
0.803 |
0.747 |
0.859 |
0.754 |
0.777 |
0.743 |
0.775 |
0.718 |
0.768 |
0.721 |
| fig|211586.9.peg.2702 |
0.713 |
0.744 |
0.788 |
0.812 |
0.742 |
0.769 |
0.780 |
0.774 |
0.811 |
0.809 |
0.837 |
0.858 |
|
0.760 |
0.751 |
0.782 |
0.803 |
0.726 |
0.717 |
0.709 |
0.722 |
0.712 |
0.746 |
0.650 |
0.747 |
0.705 |
0.667 |
0.616 |
| fig|211586.9.peg.2682 |
0.828 |
0.845 |
0.874 |
0.906 |
0.916 |
0.929 |
0.908 |
0.929 |
0.916 |
0.892 |
0.844 |
0.871 |
0.760 |
|
0.869 |
0.941 |
0.912 |
0.851 |
0.926 |
0.907 |
0.847 |
0.770 |
0.763 |
0.901 |
0.728 |
0.771 |
0.819 |
0.740 |
| fig|211586.9.peg.2680 |
0.859 |
0.856 |
0.862 |
0.872 |
0.863 |
0.877 |
0.856 |
0.850 |
0.876 |
0.867 |
0.835 |
0.815 |
0.751 |
0.869 |
|
0.869 |
0.930 |
0.847 |
0.835 |
0.825 |
0.683 |
0.872 |
0.903 |
0.762 |
0.867 |
0.737 |
0.715 |
0.642 |
| fig|211586.9.peg.2683 |
0.819 |
0.850 |
0.860 |
0.892 |
0.898 |
0.897 |
0.917 |
0.913 |
0.924 |
0.894 |
0.825 |
0.870 |
0.782 |
0.941 |
0.869 |
|
0.919 |
0.899 |
0.927 |
0.896 |
0.777 |
0.791 |
0.813 |
0.883 |
0.776 |
0.787 |
0.801 |
0.718 |
| fig|211586.9.peg.2681 |
0.852 |
0.865 |
0.894 |
0.904 |
0.883 |
0.895 |
0.884 |
0.885 |
0.915 |
0.896 |
0.864 |
0.875 |
0.803 |
0.912 |
0.930 |
0.919 |
|
0.867 |
0.868 |
0.869 |
0.763 |
0.868 |
0.877 |
0.795 |
0.852 |
0.764 |
0.738 |
0.708 |
| fig|211586.9.peg.2675 |
0.735 |
0.828 |
0.820 |
0.841 |
0.850 |
0.840 |
0.901 |
0.887 |
0.886 |
0.866 |
0.818 |
0.833 |
0.726 |
0.851 |
0.847 |
0.899 |
0.867 |
|
0.827 |
0.762 |
0.719 |
0.876 |
0.849 |
0.769 |
0.849 |
0.708 |
0.755 |
0.687 |
| fig|211586.9.peg.2689 |
0.865 |
0.826 |
0.845 |
0.857 |
0.926 |
0.905 |
0.907 |
0.909 |
0.908 |
0.895 |
0.777 |
0.803 |
0.717 |
0.926 |
0.835 |
0.927 |
0.868 |
0.827 |
|
0.931 |
0.789 |
0.743 |
0.728 |
0.926 |
0.692 |
0.816 |
0.837 |
0.794 |
| fig|211586.9.peg.2684 |
0.835 |
0.738 |
0.776 |
0.801 |
0.859 |
0.853 |
0.831 |
0.844 |
0.862 |
0.859 |
0.706 |
0.747 |
0.709 |
0.907 |
0.825 |
0.896 |
0.869 |
0.762 |
0.931 |
|
0.765 |
0.754 |
0.713 |
0.890 |
0.676 |
0.834 |
0.810 |
0.786 |
| fig|211586.9.peg.2707 |
0.680 |
0.709 |
0.765 |
0.818 |
0.812 |
0.828 |
0.814 |
0.850 |
0.822 |
0.822 |
0.782 |
0.859 |
0.722 |
0.847 |
0.683 |
0.777 |
0.763 |
0.719 |
0.789 |
0.765 |
|
0.612 |
0.575 |
0.804 |
0.563 |
0.725 |
0.826 |
0.792 |
| fig|211586.9.peg.2677 |
0.754 |
0.778 |
0.768 |
0.767 |
0.775 |
0.775 |
0.804 |
0.792 |
0.842 |
0.851 |
0.780 |
0.754 |
0.712 |
0.770 |
0.872 |
0.791 |
0.868 |
0.876 |
0.743 |
0.754 |
0.612 |
|
0.897 |
0.647 |
0.925 |
0.706 |
0.660 |
0.663 |
| fig|211586.9.peg.2676 |
0.746 |
0.760 |
0.768 |
0.771 |
0.739 |
0.743 |
0.767 |
0.748 |
0.820 |
0.798 |
0.766 |
0.777 |
0.746 |
0.763 |
0.903 |
0.813 |
0.877 |
0.849 |
0.728 |
0.713 |
0.575 |
0.897 |
|
0.655 |
0.903 |
0.661 |
0.605 |
0.546 |
| fig|211586.9.peg.2688 |
0.735 |
0.709 |
0.737 |
0.786 |
0.816 |
0.817 |
0.812 |
0.829 |
0.821 |
0.797 |
0.670 |
0.743 |
0.650 |
0.901 |
0.762 |
0.883 |
0.795 |
0.769 |
0.926 |
0.890 |
0.804 |
0.647 |
0.655 |
|
0.596 |
0.736 |
0.795 |
0.706 |
| fig|211586.9.peg.2679 |
0.715 |
0.776 |
0.763 |
0.770 |
0.732 |
0.729 |
0.778 |
0.754 |
0.821 |
0.812 |
0.795 |
0.775 |
0.747 |
0.728 |
0.867 |
0.776 |
0.852 |
0.849 |
0.692 |
0.676 |
0.563 |
0.925 |
0.903 |
0.596 |
|
0.627 |
0.568 |
0.566 |
| fig|211586.9.peg.2704 |
0.705 |
0.623 |
0.652 |
0.694 |
0.775 |
0.768 |
0.799 |
0.784 |
0.822 |
0.821 |
0.658 |
0.718 |
0.705 |
0.771 |
0.737 |
0.787 |
0.764 |
0.708 |
0.816 |
0.834 |
0.725 |
0.706 |
0.661 |
0.736 |
0.627 |
|
0.899 |
0.842 |
| fig|211586.9.peg.2705 |
0.667 |
0.639 |
0.666 |
0.732 |
0.811 |
0.804 |
0.849 |
0.833 |
0.846 |
0.830 |
0.691 |
0.768 |
0.667 |
0.819 |
0.715 |
0.801 |
0.738 |
0.755 |
0.837 |
0.810 |
0.826 |
0.660 |
0.605 |
0.795 |
0.568 |
0.899 |
|
0.857 |
| fig|211586.9.peg.2706 |
0.703 |
0.650 |
0.702 |
0.711 |
0.798 |
0.788 |
0.809 |
0.802 |
0.817 |
0.834 |
0.653 |
0.721 |
0.616 |
0.740 |
0.642 |
0.718 |
0.708 |
0.687 |
0.794 |
0.786 |
0.792 |
0.663 |
0.546 |
0.706 |
0.566 |
0.842 |
0.857 |
|
Pegs in Atomic Regulon 45 [ON=51 OFF=181]
The hypothetical stands out, does it not?
Shewnella and Microcystins??
"Microcystins are cyclic nonribosomal peptides produced by cyanobacteria. They are cyanotoxins and can be very toxic for plants and animals including humans. " (Wikipedia)
The known bacteria that are able to affect the cyanobacterial growth
include species such as Cytophaga, Shewanella, Streptomyces and Vibrio
strains (Yamamoto et al. 1998, Yoshikawa et al. 2000, Rashidan and
Bird 2001, Manage et al. 2000, Salomon et al. 2003). They can inhibit
the cyanobacterial growth by means such as competition for limiting
nutrients and by production of diffusible lytic compounds
...
cyanobacteria together with heterotrophic bacteria may represent an
increased health risk. The infection susceptibility of persons exposed
to cyanobacterial water blooms could be lowered by the adverse effects
of the cyanobacterial toxins. The lipopolysaccharides of the
Gram-negative bacteria, in turn, inhibit glutathione S-transferase
activity, which is important in detoxifying of microcystins, and may
increase the risk posed by the cyanobacterial hepatotoxins
Why Are Such Conjectures Easy to Produce?
Now, let me proceed to the second point of this discussion: the factors that make it
possible to produce hundreds of these conjectures for minimal effort.
I must begin with an overview on how atomic regulons are computed:
-
The processing begins by computing estimates of coregulated clusters. These
estimates are based on two sources:
-
estimates of co-expression based on position on
the chromosome considered in the context of a set of expression experiments, and
-
estimates of co-expression based on presence in the same row of a subsystem maintained within the SEED.
These estimates might reasonably be called the initial set of putative atomic regulons.
-
After the initial set is computed, a computation is run that tries to reconcile the raw expression
values with these assertions. That is, we seek a set of ON/OFF calls for each gene, and then for each putative
atomic regulon, that come as close as we can to the raw expression values.
- Once we have estimates for ON/OFF values for each atomic regulon in each experiment, we can compute a
profile for each atomic regulon as a vector of ON/OFF values corresponding to the experiments.
We can group merge an two putative atomic regulons with identical profiles, producing the final set of
atomic regulons.
This simple process led to formation of 483 atomic regulons for E.coli.
What is astounding is the wealth of information in just combining operon, subsystem, and expression
data. There is a great deal to be offered by other forms of information, as I will argue a bit
later in this document. However, I urge the reader not to focus on the available information that
we did not use in the previous discussion (e.g., the actual conditions used in the experiments,
metabolic modeling, or functional coupling data based on comparative analysis of many genomes); rather,
focus on how much falls out of just quickly formed conjectures (i.e., potential operons and
presence in a common subsystem) together with the expression data.
I would look to meander here to an example of how the addition of expression data to
co-occurrence data can lead to useful refinements. To understand this short aside, you
will need to briefly examine a set of PEGs in the E.coli genome. I would suggest
going to
fig|83333.1.peg.648.
If you hover over genes in this region, you will note that a SEED annotator has placed genes peg.644
through peg.652 into a single, cluster-based subsystem. The reason was almost certainly that this
large group of genes are in a fairly fixed cluster (but not an operon), and when there is no obvious "core"
of the cluster. That is, when you look through the current genomes, you see the entire cluster within
a close taxonomic group of the enteric bacteria, but the group does not extend much past that.
There seemed to be value in recording the cluster, so a subsystem was formed. However,
when we add expression data, we see the following correlation matrix:
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.644 |
Uncharacterized protein YbeL / FIG002095: hypothetical protein |
A_hypothetical_protein_that_co-occurs_with_Leucyl-tRNA_synthetase |
| fig|83333.1.peg.645 |
Uncharacterized protein YbeQ |
|
| fig|83333.1.peg.646 |
Uncharacterized protein YbeR |
|
| fig|83333.1.peg.647 |
Uncharacterized J domain-containing protein YbeS, predicted chaperone |
|
| fig|83333.1.peg.648 |
Uncharacterized protein YbeT |
|
| fig|83333.1.peg.649 |
Uncharacterized protein YbeU |
|
| fig|83333.1.peg.650 |
Uncharacterized J domain-containing protein YbeV |
|
| fig|83333.1.peg.651 |
Chaperone protein hscC (Hsc62) |
|
| fig|83333.1.peg.652 |
Pyrimidine-specific ribonucleoside hydrolase RihA (EC 3.2.-.-) |
Queuosine-Archaeosine_Biosynthesis |
This changes the picture significantly. Notice that the first and last genes in the "cluster",
peg.644 and peg.652, are clearly not related to the rest of the genes.
peg.651 is pretty weakly related. This changes one's view of the significance of the cluster.
Without the expression data, it was natural to wonder whether the group was related to the
"pyrimidine-specific hydrolase RihA"; with the expression data, this notion should probably be
discarded.
Let me show you another of these situations, lest you think my last one was an anomaly.
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.699 |
FIG143828: Hypothetical protein YbgA |
COG3380_COG2907,EC699-706 |
| fig|83333.1.peg.700 |
Deoxyribodipyrimidine photolyase (EC 4.1.99.3) |
COG3380_COG2907,DNA_repair,_bacterial_photolyase,EC699-706 |
| fig|83333.1.peg.701 |
Di/tripeptide permease YbgH |
EC699-706,Proton-dependent_Peptide_Transporters |
| fig|83333.1.peg.702 |
FIG042796: Hypothetical protein |
EC699-706 |
| fig|83333.1.peg.703 |
Allophanate hydrolase 2 subunit 1 (EC 3.5.1.54) |
EC699-706,Urea_carboxylase_and_Allophanate_hydrolase_cluster |
| fig|83333.1.peg.704 |
Allophanate hydrolase 2 subunit 2 (EC 3.5.1.54) |
EC699-706,Urea_carboxylase_and_Allophanate_hydrolase_cluster |
| fig|83333.1.peg.705 |
Lactam utilization protein LamB |
EC699-706 |
| fig|83333.1.peg.706 |
Endonuclease VIII |
DNA_Repair_Base_Excision,EC699-706 |
Here it becomes apparent that the cluster is really peg.702 through peg.705.
I believe that I was the author of the original, large cluster-based subsystem. In the presence
of the expression data, I would have focused on the smaller cluster.
Before leaving this topic and returning to broader issues, let me bring your attention
to the cluster including peg.3965 through peg.3967. You can view the cluster
here.
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.3965 |
NMN phosphatase (EC 3.1.3.5); Class B acid phosphatase precursor (EC 3.1.3.2) |
NAD_and_NADP_cofactor_biosynthesis_global |
| fig|83333.1.peg.3966 |
hypothetical protein |
|
| fig|83333.1.peg.3967 |
Protein yjbR |
|
It becomes obvious that peg.3965 is not co-expressed with the other two genes, which is quite unfortunate, since it was the gene with a function assigned to it.
I have shown a number of instances in which the information produced by the expression data was quite
suggestive. I feel that I should also present one in which I can see no pattern at all, although the
correlation coefficients are quite high:
Functions in Escherichia coli K12
| PEG |
Function |
Subsystems |
| fig|83333.1.peg.991 |
Flavoprotein wrbA |
|
| fig|83333.1.peg.1766 |
Uncharacterized protein YeaG |
Unknown_carbohydrate_utilization_(_cluster_Ydj_) |
| fig|83333.1.peg.2072 |
Fructose-bisphosphate aldolase class I (EC 4.1.2.13) |
Calvin-Benson_cycle,Formaldehyde_assimilation:_Ribulose_monophosphate_pathway,Glycolysis_and_Gluconeogenesis,Unknown_carbohydrate_utilization_(_cluster_Yeg_) |
| fig|83333.1.peg.2056 |
hypothetical protein |
|
| fig|83333.1.peg.798 |
Non-specific DNA-binding protein Dps / Iron-binding ferritin-like antioxidant protein / Ferroxidase (EC 1.16.3.1) |
Cobalt-zinc-cadmium_resistance,Oxidative_stress,YgfZ-Iron |
| fig|83333.1.peg.1174 |
FIG004684: SpoVR-like protein |
|
| fig|83333.1.peg.1881 |
Trehalose-6-phosphate phosphatase (EC 3.1.3.12) |
Trehalose_Biosynthesis |
| fig|83333.1.peg.1722 |
Osmotically inducible lipoprotein E precursor |
|
| fig|83333.1.peg.3296 |
Cell filamentation protein fic |
|
We should revisit this cluster in a few years to see whether or not
the truth is hidden as deeply as it appears.
These Conjectures are Valuable, but They are Really Only a By-Product of a Far More Important Advance
In this section, I will present the view that the conjectures that are
now emerging (and, there will easily be hundreds of them) are important, but
not the really valuable part of what is happening.
To understand the real step forward, we need to think about where we are going. I claim that the next big
step in filling in our understanding of unicellular life will be to model microbes as finite-state machines that
make transitions between states in response to a changing environment. I believe that some readers will
consider my assertion to be obvious and not worth much thought; a second set will consider it silly and obviously
counterproductive. To understand the significance of the assertion, it will be necessary to make it
completely precise. So, let me try.
Let us specifically consider what I mean in the context of Escherichia coli, the organism that I
used in the examples above. The goal will be to go through the following steps:
- Define a set of approximate atomic regulons. We have a reasonable starting point with the 483 existing
sets of genes. However, this set needs to be substantially extended. At this point 2079 genes have been placed into
these putative atomic regulons, and this amounts to less than half of the genes in E.coli.
We have made no effort to form a comprehensive set.
- Define a small set of states of the cell. Initially, this will probably be
very few. This is a somewhat arbitrary process, since the notion of "state" is
relatively arbitrary. I believe that we will move towards a basic hierarchy of states.
That is, we will have a few large states, each of which contains a number of "substates".
There is no question state, as a technical task, this is not difficult. However, to be
meaningful the set of states must reflect the same reality skilled microbial physiologists
use to organize their thoughts.
- Understand the experimental conditions that determine the atomic regulons.
To date, we have made no attempt to understand the relationship between experimental conditions and
atomic regulons. It may be prohibitively difficult to actually learn anything.
- Create plausible metabolic models for each state of the cell.
I am more enthusiastic about the use of metabolic modeling. The goal
of "Given a set of reactions corresponding to a putative state of the cell (i.e.,
harvested from the set of atomic regulons that are ON in the state), predict what extra
machinery is needed for the cell to function" is the sort of effort that will force
merging of atomic regulons and will drag in new machinery absent from the initial model.
- Force the remaining genes into approximate atomic regulons. The remaining genes not in atomic regulons
can be forced into the initial atomic regulons with the closest expression profiles, or into new atomic regulons based
on a notion of "minimal distance" between profiles.
- Define the regulatory model.
Once the states and atomic regulons have been imposed, the regulatory model can be studied.
Clearly, the notion is that we have a limited set of states, and that transitions between states occur.
As each transition is proposed and verified, the corresponding regulatory apparatus can be
characterized. Much os this is already done for E.coli, but I confess that it is not obvious
to me how the set of transitions has been defined. I believe that a great deal is known about the roles
of specific regulators, but connecting this body of work with the simplistic view based on atomic regulons
will be a substantial amount of work.
By listing some of these steps, I do not mean to say "This is what we will do." I offer them solely
as a suggestion of steps we might take; but I do hold the view that taking these simple steps
will force a radical advance in the field. By imposing a structured view of the genes, metabolism,
and regulation (in terms of "states" and "atomic regulons"), we impose the framework that allows conjectures
of the sort we saw in the first section to emerge.
The Pipeline that Needs to be Built
In the last section, I argued that we were progressing towards the goal of defining states of the cell,
atomic regulons, and rules governing transitions between states. In this section, I would like
to just make some comments about specific projects that should be started.
I would begin by saying that the very limited expression data that we have been able to gather is proving
to be extremely useful. We need to get data for more organisms, and we need at least text descriptions
of the experimental conditions. I have tried to participate in this process, but it is a tedious, complex
process that breaks down in many ways. We need to architect a protocol that will allow us to absorb
rapidly growing amounts of data in a form that is useful to us. Since the availability of expression data
make skyrocket due to new sequencing capabilities, I would sugest focusing on working with a limited
set of groups, forcing minimal consistency, and adding more organisms as large amounts of data become available.
For now, let us get a few more genomes, clean up what we have, and try to capture the algorithms used
to create the existing data.
The next step must almost certainly be picking a small number of genomes with substantial expression data
and push the attempt to form comprehensive and reasonably accurate atomic regulons. I would favor
E.coli, Bacillus subtilis, and Shewanella. Good arguments could be made for adding
strains of Clostridia, and Salmonella. We should form partnerships in each case.
In each case, a "working set" of atomic regulons should be formed. Then, each experiment should be viewed
as a "state" (or, more precisely, as a minor variant of a small set of "states"). That is, it should be possible
for the cell to grow under the conditions omost experiments, and for those experiments we should be able
to construct a metabolic network that is viable. If we cannot, we have an inconsistency, and these should be systematically
removed.
I believe that, if we push 3-5 organisms through to the point where we have a reasonable estimate of states and atomic regulons,
it will lead almost immediately to rapid advances in analysis of regulation and clarification of the functions of
the remaining "hypothetical genes".


